There's a slow-burning discussion on taxonomic concepts on Github that I am half participating in. As seems inevitable in any discussion of taxonomy, there's a lot of floundering about given that there's lots of jargon - much of it used in different ways by different people - and people are coming at the problem from different perspectives.
In one sense, taxonomy is pretty straightforward. We have taxonomic names (labels), we have taxa (sets) that we apply those labels to, and a classification (typically a set of nested sets, i.e., a tree) of those taxa. So, if we download, say, GenBank, or GBIF, or BOLD we can pretty easily model names (e.g., a list of strings), the taxonomic tree (e.g., a parent-child hierarchy), and we have a straightforward definition of the terminal taxa (leaves) or the tree: they comprise the specimens and observations (GBIF), or sequences (GenBank and BOLD) assigned to that taxon (i.e., for each specimen or sequence we have a pointer to the taxon to which it belongs).
Given this, one response to the taxonomic concept discussion is to simply ignore it as irrelevant, and we can demonstrably do a lot of science without it. I suspect most people dealing with GBIF and GenBank data aren't aware of the taxonomic concept issue. Which begs the question, why the ongoing discussion about concepts?
Perhaps the fundamental issue is that taxonomic classification changes over time, and hence the interpretation of a taxon can change over time. In other words, the problem is one of versioning. Once again, the simplest strategy to deal with this is simply use the latest version. In much the same way that most of us probably just read the latest version of a Wikipedia page, and many of us are happy to have our phone apps update automatically, I suspect most are happy to just grab the latest version and do some, you know, science.
I think taxonomic concepts really become relevant when we are aggregating data from sources where the data may not be current. In other words, where data is associated with a particular taxonomic name and the interpretation of that name has changed since the last time the data was curated. If the relationships of a taxon or specimen can be computed on the fly, e.g. if the data is a DNA barcode, then this issue is less relevant because we can simply re-cluster the sequences and discover where the specimen with that sequence belongs in a new classification. But for many specimens we don't have sufficient information to do this computation (this is one reason DNA barcodes are so useful, everything needed to determine a barcode's relationship is contained in the sequence itself).
To make this concrete, consider the genus Brookesia in GBIF (GBIF:2449310.
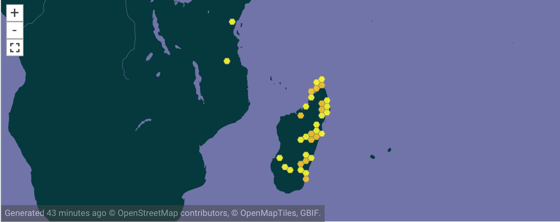
According to Wikipedia Brookesia is endemic to Madagascar, so why does it appear on the African mainland? There are two records from Africa, Brookesia brookesia ionidesi collected in 1957 and Brookesia temporalis collected in 1926. Both represent taxa that were in the genus Brookesia at one point, but are now in different genera. So our notion of Brookesia has changed over time, but curation of these records has yet to catch up with that.
So, what would be ideal would be if we have a timestamped series of classifications so that we could go back in time and see what a given taxon meant at a given time, and then go forward to see the status of that taxon today. Having such a timestamped series is not a trivial task, indeed it may only be available in well studied groups. Birds are one such group, where each year eBird updates the current bird classification based on taxonomic activity over the previous year. As part of the Github discussion I posted visual "diff" between two bird classifications:

You can see the complete diff here, and the blog post Visualising the difference between two taxonomic classifications for details on the method.. The illustration above shows the movement of one species from Sasia to Verreauxia.
So, given two classifications we can compute the difference between them, and represent that difference as an "edit script" or operations to convert one tree into another. These edits are essentially what taxonomists do when they revise a group, they do things such as move species form one genus to another, merge some taxa, sink others into synonymy, and so on. So, taxonomy is essentially creating a series of edit files ("patches") to a classification. At a recent workshop in Ottawa Karen Cranston pointed out that the Open Tree of Life has been accumulating amendments to their classification and that these are essentially patch files.
Hence, we could have a markup language for taxonomic work that described that work in terms of edit operations that can then be automatically applied to an existing classification. We could imagine encoding all the bird taxonomy for a year in this way, applying those patches to the previous years' tree, and out pops the new classification. The classification becomes an evolving document under version control (think GitHub for trees). Of course, we'd need something to detect whether two different papers were proposing incompatible changes, but that's essentially a tree compatibility problem.
One way to store version information would be to use time-based versioned graphs. Essentially, we start with each node in the classification tree having a start date (e.g., 2017) and an open-ended end date. A taxonomic work post 2017 that, say, moved a species from one genus to another would set the end date for the parent-child link between genus and species, and create a new timestamped node linking the species to its new genus. To generate the 2018 classification we simply extract all links in the tree whose date range includes 2018 (which means the old generic assignment for the species is not included). This approach gives us a mechanism for automating the updating of a classification, as well as time-based versioning.
I think something along these lines would create something useful, and focus the taxonomic discussion on solving a specific problem.