OK, so the title is pure click bait, but here's the thing. It seems to me that the Semantic Web as classically conceived (RDF/XML, SPARQL, triple stores) has had relatively little impact outside academia, whereas other technologies such as JSON, NoSQL (e.g., MongoDB, CouchDB) and graph databases (e.g., Neo4J) have got a lot of developer mindshare.
In biodiversity informatics the Semantic Web has been a round for a while. We've been pumping out millions of RDF documents (mostly served by LSIDs) since 2005 and, to a first approximation, nothing has happened. I've repeatedly blogged about why I think this is (see this post for a summary).
I was an early fan of RDF and the Semantic Web, but soon decided that it was far more hassle than it was worth. The obsession with ontologies, the problems of globally unique identifiers based on HTTP (http-14 range, anyone?), the need to get a lot of ducks in a row all mad it a colossal pain. Then I discovered the NoSQL document database CouchDB, which is a JSON store that features map-reduce views rather than on the fly queries. To somebody with a relational database background this is a bit of a headfuck:

But CouchDB has a great interface, can be replicated to the cloud, and is FUN (how many times can you say that about a database?). So I starting playing with CouchDB for small projects, then used it to build BioNames and more recently moved BioStor to CouchDB hosted both locally and in the cloud.
Then there are graph databases such as Neo4J, which has some really cool things such as GraphGists which is a playground where you can create interactive graphs and query them (here's an example I created). Once again, this is FUN.
Another big trend over the last decade is the flight from XML and its hideous complexities (albeit coupled with great power) to the simplicity of JSON (part of the rise of JavaScript). JSON makes it very easy to pass around data in simple key-value documents (with more complexity such as lists if you need them). Pretty much any modern API will serve you data in JSON.
So, what happened to RDF? Well, along with a plethora of formats (XML, triples, quads, etc., etc.) it adopted JSON in the form of JSON-LD (see JSON-LD and Why I Hate the Semantic Web for background). JSON-LD lets you have data in JSON (which both people and machines find easy to understand) and all the complexity/clarity of having the data clearly labelled using controlled vocabularies such as Dublin Core and schema.org. This complexity is shunted off into a "@context" variable where it can in many cases be safely ignored.
But what I find really interesting is that instead of JSON-LD being a way to get developers interested in the rest of the Semantic Web stack (e.g. HTTP URIs as identifiers, SPARQL, and triple stores), it seems that what it is really going to do is enable well-described structured to get access to all the cool things being developed around JSON. For example, we have document databases such as CouchDB which speaks HTTP and JSON, and search servers such as ElasticSearch which make it easy to work with large datasets. There are also some cool things happening with graph databases and Javascript, such as Hexastore (see also Weiss, C., Karras, P., & Bernstein, A. (2008, August 1). Hexastore. Proc. VLDB Endow. VLDB Endowment. http://doi.org/10.14778/1453856.1453965, PDF here) where we create the six possible indexes of the classic RDF [subject,predicate,object] triple (this is the sort of thing can also be done in CouchDB). Hence we can have graph databases implemented in a web browser!
So, when we see large-scale "Semantic Web" applications that actually exist and solve real problems, we may well be more likely to see technologies other than the classic Semantic Web stack. As an example, see the following paper:
Szekely, P., Knoblock, C. A., Slepicka, J., Philpot, A., Singh, A., Yin, C., … Ferreira, L. (2015). Building and Using a Knowledge Graph to Combat Human Trafficking. The Semantic Web - ISWC 2015. Springer Science + Business Media. http://doi.org/10.1007/978-3-319-25010-6_12
There's a free PDF here, and a talk online. The consortium behind this project researchers did extensive text mining, data cleaning and linking, creating a massive collection of JSON-LD documents. Rather than use a triple store and SPARQL, they indexed the JSON-LD using ElasticSearch (notice that they generated graphs for each of the entities they care about, in a sense denormalising the data).
I think this is likely to be the direction many large-scale projects are going to be going. Use the Semantic Web ideas of explicit vocabularies with HTTP URIs for definitions, encode the data in JSON-LD so it's readable by developers (no developers, no projects), then use some of the powerful (and fun) technologies that have been developed around semi-structured data. And if you have JSON-LD, then you get SEO for free by embedding that JSON-LD in your web pages.
In summary, if biodiversity informatics wants to play with the Semantic Web/linked data then it seems obvious that some combination of JSON-LD with NoSQL, graph databases, and search tools like ElasticSearch are the way to go.

 This guest post by
This guest post by  Figure 1 shows a herbarium specimen, but there's no identifier or link for the specimen. Is this specimen available online? Can I see it in GBIF? Can I see it's history and explore further? if not, why not? If it's not available online, why not pick one that is?
Figure 1 shows a herbarium specimen, but there's no identifier or link for the specimen. Is this specimen available online? Can I see it in GBIF? Can I see it's history and explore further? if not, why not? If it's not available online, why not pick one that is?
 Google Maps uses "tiles" to create a zoomable interface, so we need to create tiles for different zoom levels for the phylogeny. To create this visualisation I did the following:
Google Maps uses "tiles" to create a zoomable interface, so we need to create tiles for different zoom levels for the phylogeny. To create this visualisation I did the following:


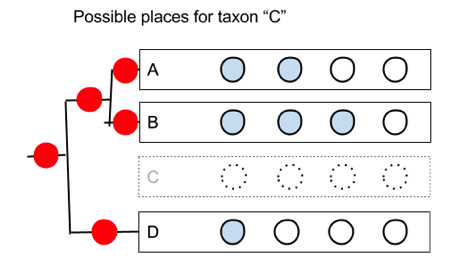 But if we simply shuffle the order of the taxa we can’t generate all the trees. However, if we remember that we also have the internal nodes, then there is a simple way we can generate the trees.
But if we simply shuffle the order of the taxa we can’t generate all the trees. However, if we remember that we also have the internal nodes, then there is a simple way we can generate the trees.  When we draw a tree each row corresponds to a node. The gap between each pair of leaves (the taxa A,B,D) corresponds to the an internal nodes. So we could divide the drawing up into “hit zone”, so that if you drag the taxon we’re adding (“C”) onto the zone centred on a leaf, we add the taxon below that leaf; if we drag it onto a zone between two leaves, we attach it below that the corresponding internal node. From the user’s point of view they are still simply sliding taxa up and down, but in doing so we can create each of the possible trees.
When we draw a tree each row corresponds to a node. The gap between each pair of leaves (the taxa A,B,D) corresponds to the an internal nodes. So we could divide the drawing up into “hit zone”, so that if you drag the taxon we’re adding (“C”) onto the zone centred on a leaf, we add the taxon below that leaf; if we drag it onto a zone between two leaves, we attach it below that the corresponding internal node. From the user’s point of view they are still simply sliding taxa up and down, but in doing so we can create each of the possible trees.
 Google knows how many species there are. More significantly, it knows what I mean when I type in "how many species are there". Wouldn't it be nice to be able to do this with biodiversity databases? For example, how many species of insect are found in Fiji? How would you answer this question? I guess you'd Google it, looking for a paper. Or you'd look in vain on GBIF, and then end up hacking some API queries to process data and come up with an estimate. Why can't we just ask?
Google knows how many species there are. More significantly, it knows what I mean when I type in "how many species are there". Wouldn't it be nice to be able to do this with biodiversity databases? For example, how many species of insect are found in Fiji? How would you answer this question? I guess you'd Google it, looking for a paper. Or you'd look in vain on GBIF, and then end up hacking some API queries to process data and come up with an estimate. Why can't we just ask? All very sensible questions that existing biodiversity databases would struggle to answer.
All very sensible questions that existing biodiversity databases would struggle to answer. Over the weekend, out of the blue, Dan Whaley commented on an earlier blog post of mine (
Over the weekend, out of the blue, Dan Whaley commented on an earlier blog post of mine (
 A little over a week ago I was at the
A little over a week ago I was at the  I need more time to sketch this out fully, but I think a case can be made for a taxonomy-centric (or, perhaps more usefully, a biodiversity-centric) clone of PubMed Central.
I need more time to sketch this out fully, but I think a case can be made for a taxonomy-centric (or, perhaps more usefully, a biodiversity-centric) clone of PubMed Central. The page image only appears if you click on the blue labels for the page. None of this is robust or optimised, but it is a workable proof-of-concept of how fill-text search could work.
The page image only appears if you click on the blue labels for the page. None of this is robust or optimised, but it is a workable proof-of-concept of how fill-text search could work. One of the less glamorous but necessary tasks of data cleaning is mapping "strings to things", that is, taking strings such as "George A. Boulenger" and mapping them to identifiers, such as
One of the less glamorous but necessary tasks of data cleaning is mapping "strings to things", that is, taking strings such as "George A. Boulenger" and mapping them to identifiers, such as  Imagine a web site where researchers can go, log in (easily) and get a list of all the species they have described (with pretty pictures and, say, GBIF map), and a list of all DNA sequences/barcodes (if any) that they've published. Imagine that this is displayed in a colourful way (e.g., badges), and the results tweeted with the hastag #itaxonomist.
Imagine a web site where researchers can go, log in (easily) and get a list of all the species they have described (with pretty pictures and, say, GBIF map), and a list of all DNA sequences/barcodes (if any) that they've published. Imagine that this is displayed in a colourful way (e.g., badges), and the results tweeted with the hastag #itaxonomist.


 In my (
In my (
