Currently in classes where I teach the basics of tree building, we still fire up ancient iMacs, load up MacClade, and let the students have a play. Typically we give them the same data set and have a class competition to see which group can get the shortest tree by manually rearranging the branches. It’s fun, but the computers are old, and what’s nostalgic for me seems alien to the iPhone generation.
One thing I’ve always wanted to have is a simple MacClade-like tree editor for the Web, where the aim is not so much character analysis as teaching the basics of tree building. Something with the easy of use as Phylo (basically Candy Crush for sequence alignments).

The challenge is to keep things as simple as possible. One idea is to have a column of taxa and you can drag individual taxa up and down to rearrange the tree.

Imagine each row has the characters and their states. Unlike the Phylo game, where the goal is to slide the amino acids until you get a good aliognment, here we want to move the taxa to improve the tree (e.g., based on its parsimony score).
The problem is that we need to be able to generate all possible rearrangements for a given number of taxa. In the example above, if we move taxon C, there are five possible positions it could go on the remaining subtree:
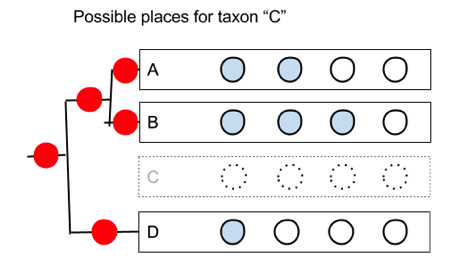

We could implement this in a Web browser with some Javascript to handle the user moving the taxa, and draw the corresponding phylogeny to the left, and quickly update the (say, parsimony) score of the tree so that the user gets immediate feedback as to whether the rearrangement they’ve made improves the tree.
I think this could be a fun teaching tool, and if it supported touch then students could use their phones and tablets to get a sense of how tree building works.