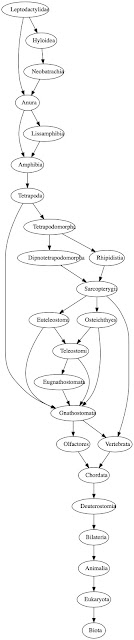
Classifications in Wikidata can be complex and are often not trees. For example, if we trace the parents of the frog family Leptodactylidae back we get a graph like this:
Each oval represents a taxon in Wikidata, and each arrow connects a taxon to its parent(s) in Wikidata. Likewise, if we do the same for the albatross genus Diomedea we get a similarly complex diagram:
The presence of multiple classifications likely reflects several factors. If you deal with just extant species you are likely to have fairly shallow classifications, for example, the kingdom, phylum, class, order, family, genus ranks used by GBIF. may be enough. Some taxonomic groups may routinely use ranks such as subfamily, and in well-studied groups there may be additional taxa based on phylogenetic research (e.g., the RTA clade in spiders). And of course, different Wikidata editors may favour different classifications.
Anecdotally (certainly for vertebrates), many of the additional levels in the classifications in Wikidata come from fossil taxa. In the case of birds, extant Aves (birds) are a fairly isolated group in the tree of life, but as we go down the tree towards their common ancestor with the crocodilians we encounter dinosaurs and other taxa. So if you are a palaeontologist the jump from, say Aves to Tetrapoda skips over a fairly significant part of the tree!
Faced with this complexity, how do we display a Wikidata classification in a simple way? One approach may be to display only a classification from a particular source, for example Mammal Species of the World. This requires that Wikidata has that classification, and enough information for you to extract it by a SPARQL query (for example if each node in the classification that is in MSW has a reference to MSW attached to that node).
Another approach is to extract a simplified classification from the sort of graphs shown above. Technically, these graphs are DAGs (Directed acyclic graphs). An obvious way to simplify a DAG is to find the shortest path in that DAG. For example, the path (Eukaryota, Animalia, Bilateria, Deuterostomia, Chordata, Olfactores, Gnathostomata, Tetrapoda, Amphibia, Anura, Leptodactylidae) is a path through the DAG shown above. Shortest paths are reasonably easy to find once you have a topological sorting of the graph (see e.g. Shortest Path in Directed Acyclic Graph). At the moment this looks the best bet for displaying classifications from Wikidata.
Preferred classifications
In some cases the classification in Wikidata is complicated, but this complexity isn’t reflected in SPARQL results because parts of that classification have different “ranks”. For example, for the plant order Fagales there are currently seven parents:- fabids
- Rosanae
- Hamamelididae
- eurosids I
- Monochlamydeae
- Archichlamydeae
- Juglandanae