 Following on from the previous post on putting GBIF data onto Google Maps, I'm now going to put DNA barcodes onto Google Maps. You can see the result at http://iphylo.org/~rpage/bold-map/, which displays around 1.2 million barcodes obtained from the International Barcode of Life Project (iBOL) releases. Let me describe how I made it.
Following on from the previous post on putting GBIF data onto Google Maps, I'm now going to put DNA barcodes onto Google Maps. You can see the result at http://iphylo.org/~rpage/bold-map/, which displays around 1.2 million barcodes obtained from the International Barcode of Life Project (iBOL) releases. Let me describe how I made it.Tiles
Typically when people put markers on a Google Map it is done in Javascript using the Google Maps API, and all the work is done by the browser. This works well if you haven't got too many points, but once you have more than, say, a few hundred, the browser struggles to cope. Hence, for something like GBIF or DNA barcodes where we have millions of records we need a different approach. In the GBIF data example I discussed previously I used tiles supplied by GBIF. Tiles are the basis of the "slippy maps" used by Google and others to create the experience of beig able to zoom in and out at any point on the map. At any time, the map you see in the web browser is made up of a small number of images ("tiles") that are typically 256 × 256 pixels in size. At the lowest zoom level (0) the world is represented by a single tile:

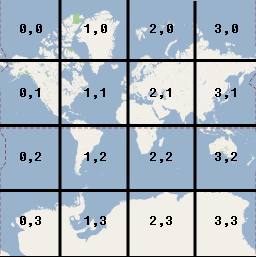
If you zoom in to the next level (1), the world now covers 41=4 tiles, zoom in again and the world now covers 42 = 16 tiles, and so on.
At each zoom level the tiles cover a smaller part of the world, and have increasing detail, so the user has the experience of zooming in closer and closer to an ever larger and more detailed map. But the browser only ever has to display enough 256 × 256 pixel tiles to fill the browser window.
Not only can we have tiles for the map of the world, we can also have tiles for data that we want to display on that map. For example, GBIF can create tiles for the hundreds of millions of occurrences in its database, so what looks like a giant map of millions of records is actually just a set of 256 x 256 tiles coloured to represent the number of records at each position on the tile.
DNA Barcodes
I wanted to make a map for DNA barcodes, but unfortunately there aren't any tiles I can use to create the map. So, I set about making my own. Here's what I did. Firstly, I downloaded the DNA barcode data from the BOLD site, and put the barcodes into a CouchDB database hosted by Cloudant. I simply parsed the tab-delimited data files and created a JSON document for each barcode, and stored that in CouchDB.
I then created a view in CouchDB that generated data for each tile. Each zoom level has its own tiles (for zoom level n there are 4n tiles). There's a nice web page on the Open Street Map wiki that describes how to compute slippy map tilenames. Here's the code I use to generate the CouchDB view:
function(doc) {
var tile_size = 256;
var pixels = 4;
if (doc.lat && doc.lon) {
for (var zoom = 0; zoom < 7; zoom++) {
var x_pos = (parseFloat(doc.lon) + 180)/360
* Math.pow(2, zoom);
var x = Math.floor(x_pos);
var relative_x = Math.round(tile_size * (x_pos - x));
var y_pos = (1-Math.log(Math.tan(parseFloat (doc.lat)
* Math.PI/180) + 1/Math.cos(parseFloat(doc.lat)
* Math.PI/180))/Math.PI)/2
* Math.pow(2,zoom);
var y = Math.floor(y_pos);
var relative_y = Math.round(tile_size * (y_pos - y));
relative_x = Math.floor(relative_x / pixels) * pixels;
relative_y = Math.floor(relative_y / pixels) * pixels;
var tile = [];
tile.push(zoom);
tile.push(x);
tile.push(y);
tile.push(relative_x);
tile.push(relative_y);
emit(tile, 1);
}
}
}
For each zoom level that I support (0 to 6) I convert the latitude and longitude of the DNA barcode sample into the coordinates of the corresponding 256 × 256 tile. I then compute the position of the sample within that tile. This is rounded to the nearest 4 pixels, which is the size of marker I've chosen to display. As an example, the barcode AMSF292-10.COI-5P has location latitude -77.8064, longitude 177.174, which for a zoom level of 6 places it in tile [63,54].
To display the marker I also need to know where in the 256 × 256 tile I should draw the marker. Coordinates in tiles start at the top left corner:

For the example above, the marker would be at x = 124, y = 200. This means that this barcode would have a key of [6, 63, 54, 124, 200] in the CouchDB view computed above. If we ignore the position within the tile, then this barcode belongs on tile [6, 63, 54].
To display the barcodes I added a layer to the Google Map. Whenever a map tile is drawn by Google maps, it calls a simple web service I created, and sends to that service the zoom level and tile coordinates for the tile it wants to draw. I then lookup that key [zoom, x, y] in CouchDB, and return all the points within that tile as a 256 x 256 SVG image. Google Maps then draws that over its own tile, and as a result the user sees both the Google Map and the location of the barcodes. To keep things manageable I only generate tiles down to zoom level 6. After that, the barcodes simply disappear.
So, with some fairly trivial coding, I've created a map tile server in CouchDB that displays over a million barcodes in Google Maps.

Hit testing
Of course, a map by itself is a bit boring. What you want to do is be able to click on a point and get some more information. If you are using the Google Maps API to add markers, then it's pretty easy to handle user clicks and do something with them. But because I'm using tiles I can't use that approach. Instead what I do is capture any clicks on the map, convert that click to tile coordinates, then query CouchDB to see if any barcodes full within that location. If so, I display them down the right side of the map. It's a bit finicky about where you click on the map, but seems to work. It would be fun to extend this approach to the GBIF map, so that you could click on a point and see what GBIF says is there.
Summary
This is all a bit crude, but as far as I'm aware this is the only interactive map of all DNA barcodes (at least, the publicly available animal barcodes). There's a lot more that could be done with this, but for now it's functional. Take it for a spin at http://iphylo.org/~rpage/bold-map/, I'd welcome any comments. If you are curious about the technical details, the source code is on GitHub at https://github.com/rdmpage/bold-map.