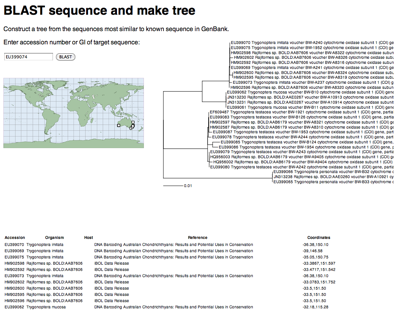
The example below is for the sequence EU399074, which falls in a cluster of "dark taxa"; in this case, DNA barcode sequences that haven't been properly labelled.
Rants, raves (and occasionally considered opinions) on phyloinformatics, taxonomy, and biodiversity informatics. For more ranty and less considered opinions, see my Twitter feed.
ISSN 2051-8188. Written content on this site is licensed under a Creative Commons Attribution 4.0 International license.
