I also ran a short, not terribly successful exercise using iTaxon to demo what semantic wikis can do. As is often the case with something that hasn't been polished yet, the students found the mechanics of doing things less than intuitive. I need to do a lot of work making data input easier (to date I've focussed on automated adding of data, and forms to edit existing data). Adding data is easy if you know how, but the user needs to know more than they really should have to.
The exercise was to take some frog taxa from the Frost et al. amphibian tree (doi:10.1206/0003-0090(2006)297[0001:TATOL]2.0.CO;2) and link them to GenBank sequences and museum specimens. The hope was that by making these links new information would emerge. You could think of it as an editable version of this. With a bit of post-exercise tidying, we got someway there. The wiki page for the Frost et al.
paper now shows a list of sequences from that paper (not all, I hasten to add), and a map for those sequences that the students added to the wiki:
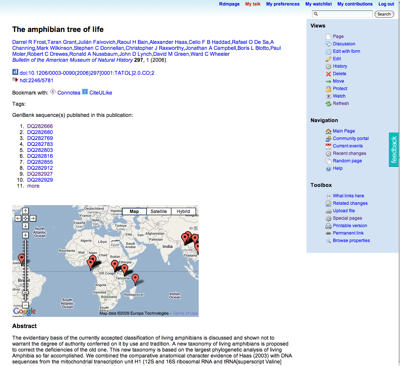
Although much remains to be done, I can't help thinking that this approach would work well for a database like TreeBASE, where one really needs to add a lot of annotation to make it useful (for example, mapping OTUs to taxon names, linking data to sequences and specimens). So, one of the things I'm going to look at is dumping a copy of TreeBASE (complete with trees) into the wiki and seeing what can be done with it. Oh, and I need to make it much, much easier for people to add data.