Triebel, D., Hagedorn, G., & Rambold, G. (2012). An appraisal of megascience platforms for biodiversity information. MycoKeys, 5(0), 45–63. doi:10.3897/mycokeys.5.4302
This paper contains a diagram that seems innocuous enough but which I find worrying:
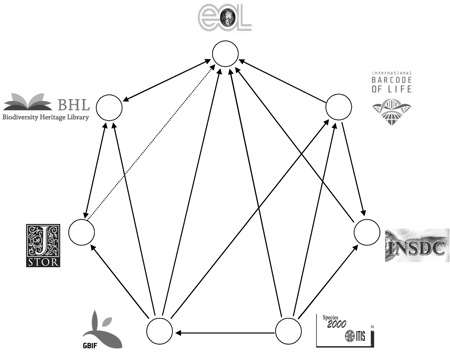
The nodes in the graph are "biodiversity megascience platforms", the edges are "cross-linkages and data exchange". What bothers me is that if you view biodiversity informatics through this lens then the relationships among these projects becomes the focus. Not the data, not the users, nor the questions we are trying to tackle. It is all about relationships between projects.
I want a different view of the landscape. For example, below is a very crude graph of the kinds of things I think about, namely kinds of data and their interrelationship:

What tends to happen is that this data landscape gets carved up by different projects, so we get separate databases of taxonomic names, images, publications, and specimens (these are the "megascience platforms" such as CoL, EOL, GBIF). This takes care of the nodes, but what about the edges, the links between the data? Typically what happens is lots of energy is expended on what to call these links, in other words, the development of the vocabularies and ontologies such as those curated by TDWG. This is all valuable work, but this doesn't tackle what for me is the real obstacle to progress, which is creating the links themselves. Where are the "megascience platforms" devoted to linking stuff together?
When we do have links between different kinds of data these tend to be within databases. For example, Genbank explicitly links sequences to publications in PubMed, and taxa in the NCBI taxonomy database. All three (sequence, publication, taxon) have identifiers (accession number, PubMed id, taxon id, respectively) that are widely used outside GenBank (and, indeed, are the de facto identifiers for the bioinformatics community). Part of the reason these identifiers are so widely used is because GenBank is the only real "megascience platform" in the list studied by Triebel et al. It's the only one that we can readily do science with (think BLAST searches, think of the number of databases that have repurposed GenBank data, or build on NCBI services).
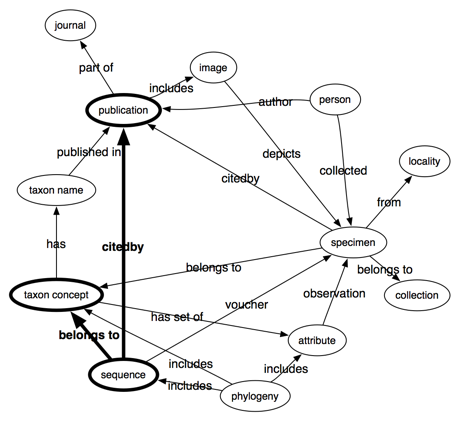
Many of the questions we might ask can be formulated as paths through a diagram like the one above. For example, if I want to do phylogeography, then I want the path phylogeny -> sequence -> specimen -> locality. If I'm lucky the phylogeny is in a database and all the sequences have been georeferenced, but often the phylogeny isn't readily available digitally, I need to map the OTUs in the tree to sequences, I then need to track down the vouchers for those sequences, and obtain the localities for those sequences from, say, GBIF. Each step involves some degree of pain as we try and map identifiers from one database to those in another.

If I want to do classical alpha taxonomy I need information on taxonomic names, concepts, publications, attributes, and specimens. The digital links between these are tenuous at best (where are the links between GBIF specimen records and the publications that cite those specimens, for example?).

Focussing on so-called "platforms" is unfortunate, in my opinion, because it means that we focus on data and how we carve up responsibility for managing it (never mind what happens to data that lacks an obvious constituency). The platforms aren't what we should be focussing on, it is the relationships between data (and no, these are not the same as the relationships between the "platforms").
If I'd like to see one thing in biodiversity informatics in 2013 it is the emergence of a "platform" that makes the links the centre of their efforts. Because without the links we are not building "platforms", we are building silos.