 Glasgow University's Institute of Biodiversity, Animal Health & Comparative Medicine, where I'm based, hosts Naturally Speaking featuring "cutting edge research and ecology banter". Apparently, what I do falls into that category, so Episode 65 features my work, specifically my entry for the 2018 GBIF Challenge (Ozymandias). The episode page has a wonderful illustration by Eleni Christoforou which captures the idea of linking things together very nicely. Making the podcast was great fun, thanks to the hosts Kirsty McWhinnie and Taya Forde. Let's face it, what academic doesn't love to talk about their own work, given half a chance? I confess I'm happy to talk about my work, but I haven't had the courage yet to listen to the podcast.
Glasgow University's Institute of Biodiversity, Animal Health & Comparative Medicine, where I'm based, hosts Naturally Speaking featuring "cutting edge research and ecology banter". Apparently, what I do falls into that category, so Episode 65 features my work, specifically my entry for the 2018 GBIF Challenge (Ozymandias). The episode page has a wonderful illustration by Eleni Christoforou which captures the idea of linking things together very nicely. Making the podcast was great fun, thanks to the hosts Kirsty McWhinnie and Taya Forde. Let's face it, what academic doesn't love to talk about their own work, given half a chance? I confess I'm happy to talk about my work, but I haven't had the courage yet to listen to the podcast.
Rants, raves (and occasionally considered opinions) on phyloinformatics, taxonomy, and biodiversity informatics. For more ranty and less considered opinions, see my Twitter feed.
ISSN 2051-8188. Written content on this site is licensed under a Creative Commons Attribution 4.0 International license.
Wednesday, December 05, 2018
Ozymandias: A biodiversity knowledge graph available as a preprint on Biorxiv
 I've written up my entry for the 2018 GBIF Challenge ("Ozymandias") and posted a preprint on Biorxiv (https://www.biorxiv.org/content/early/2018/12/04/485854). The DOI is https://doi.org/10.1101/485854 which, last time I checked, still needs to be registered.
I've written up my entry for the 2018 GBIF Challenge ("Ozymandias") and posted a preprint on Biorxiv (https://www.biorxiv.org/content/early/2018/12/04/485854). The DOI is https://doi.org/10.1101/485854 which, last time I checked, still needs to be registered.
The abstract appears below. I'll let the preprint sit there for a little while before I summon the enthusiasm to revisit it, tidy it up, and submit it for publication.
Enormous quantities of biodiversity data are being made available online, but much of this data remains isolated in their own silos. One approach to breaking these silos is to map local, often database-specific identifiers to shared global identifiers. This mapping can then be used to con-struct a knowledge graph, where entities such as taxa, publications, people, places, specimens, sequences, and institutions are all part of a single, shared knowledge space. Motivated by the 2018 GBIF Ebbe Nielsen Challenge I explore the feasibility of constructing a "biodiversity knowledge graph" for the Australian fauna. These steps involved in constructing the graph are described, and examples its application are discussed. A web interface to the knowledge graph (called "Ozymandias") is available at https://ozymandias-demo.herokuapp.com.
Thursday, November 15, 2018
Geocoding genomic databases using GBIF
 I've put a short note up on bioRxiv about ways to geocode nucleotide sequences in databases such as GenBank. The preprint is "Geocoding genomic databases using GBIF" https://doi.org/10.1101/469650.
I've put a short note up on bioRxiv about ways to geocode nucleotide sequences in databases such as GenBank. The preprint is "Geocoding genomic databases using GBIF" https://doi.org/10.1101/469650.
It briefly discusses using GBIF as a gazetteer (see https://lyrical-money.glitch.me for a demo) to geocode sequences, as well as other approaches such as specimen matching (see also Nicky Nicolson's cool work "Specimens as Research Objects: Reconciliation across Distributed Repositories to Enable Metadata Propagation" https://doi.org/10.6084/m9.figshare.7327325.v1).
Hope to revisit this topic at some point, for now this preprint is a bit of a placeholder to remind me of what needs to be done.
Thursday, October 25, 2018
Taxonomic publications as patch files and the notion of taxonomic concepts
There's a slow-burning discussion on taxonomic concepts on Github that I am half participating in. As seems inevitable in any discussion of taxonomy, there's a lot of floundering about given that there's lots of jargon - much of it used in different ways by different people - and people are coming at the problem from different perspectives.
In one sense, taxonomy is pretty straightforward. We have taxonomic names (labels), we have taxa (sets) that we apply those labels to, and a classification (typically a set of nested sets, i.e., a tree) of those taxa. So, if we download, say, GenBank, or GBIF, or BOLD we can pretty easily model names (e.g., a list of strings), the taxonomic tree (e.g., a parent-child hierarchy), and we have a straightforward definition of the terminal taxa (leaves) or the tree: they comprise the specimens and observations (GBIF), or sequences (GenBank and BOLD) assigned to that taxon (i.e., for each specimen or sequence we have a pointer to the taxon to which it belongs).
Given this, one response to the taxonomic concept discussion is to simply ignore it as irrelevant, and we can demonstrably do a lot of science without it. I suspect most people dealing with GBIF and GenBank data aren't aware of the taxonomic concept issue. Which begs the question, why the ongoing discussion about concepts?
Perhaps the fundamental issue is that taxonomic classification changes over time, and hence the interpretation of a taxon can change over time. In other words, the problem is one of versioning. Once again, the simplest strategy to deal with this is simply use the latest version. In much the same way that most of us probably just read the latest version of a Wikipedia page, and many of us are happy to have our phone apps update automatically, I suspect most are happy to just grab the latest version and do some, you know, science.
I think taxonomic concepts really become relevant when we are aggregating data from sources where the data may not be current. In other words, where data is associated with a particular taxonomic name and the interpretation of that name has changed since the last time the data was curated. If the relationships of a taxon or specimen can be computed on the fly, e.g. if the data is a DNA barcode, then this issue is less relevant because we can simply re-cluster the sequences and discover where the specimen with that sequence belongs in a new classification. But for many specimens we don't have sufficient information to do this computation (this is one reason DNA barcodes are so useful, everything needed to determine a barcode's relationship is contained in the sequence itself).
To make this concrete, consider the genus Brookesia in GBIF (GBIF:2449310.
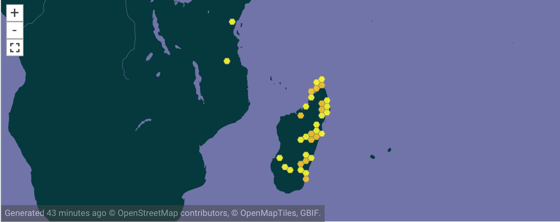
According to Wikipedia Brookesia is endemic to Madagascar, so why does it appear on the African mainland? There are two records from Africa, Brookesia brookesia ionidesi collected in 1957 and Brookesia temporalis collected in 1926. Both represent taxa that were in the genus Brookesia at one point, but are now in different genera. So our notion of Brookesia has changed over time, but curation of these records has yet to catch up with that.
So, what would be ideal would be if we have a timestamped series of classifications so that we could go back in time and see what a given taxon meant at a given time, and then go forward to see the status of that taxon today. Having such a timestamped series is not a trivial task, indeed it may only be available in well studied groups. Birds are one such group, where each year eBird updates the current bird classification based on taxonomic activity over the previous year. As part of the Github discussion I posted visual "diff" between two bird classifications:

You can see the complete diff here, and the blog post Visualising the difference between two taxonomic classifications for details on the method.. The illustration above shows the movement of one species from Sasia to Verreauxia.
So, given two classifications we can compute the difference between them, and represent that difference as an "edit script" or operations to convert one tree into another. These edits are essentially what taxonomists do when they revise a group, they do things such as move species form one genus to another, merge some taxa, sink others into synonymy, and so on. So, taxonomy is essentially creating a series of edit files ("patches") to a classification. At a recent workshop in Ottawa Karen Cranston pointed out that the Open Tree of Life has been accumulating amendments to their classification and that these are essentially patch files.
Hence, we could have a markup language for taxonomic work that described that work in terms of edit operations that can then be automatically applied to an existing classification. We could imagine encoding all the bird taxonomy for a year in this way, applying those patches to the previous years' tree, and out pops the new classification. The classification becomes an evolving document under version control (think GitHub for trees). Of course, we'd need something to detect whether two different papers were proposing incompatible changes, but that's essentially a tree compatibility problem.
One way to store version information would be to use time-based versioned graphs. Essentially, we start with each node in the classification tree having a start date (e.g., 2017) and an open-ended end date. A taxonomic work post 2017 that, say, moved a species from one genus to another would set the end date for the parent-child link between genus and species, and create a new timestamped node linking the species to its new genus. To generate the 2018 classification we simply extract all links in the tree whose date range includes 2018 (which means the old generic assignment for the species is not included). This approach gives us a mechanism for automating the updating of a classification, as well as time-based versioning.
I think something along these lines would create something useful, and focus the taxonomic discussion on solving a specific problem.
Wednesday, October 24, 2018
Specimens, collections, researchers, and publications: towards social and citation graphs for natural history collections
Being in Ottawa last week for a hackathon meant I could catch up with David Shorthouse (@dpsSpiders. David has been doing some neat work on linking specimens to identifiers for researchers, such as ORCIDs, and tracking citations of specimens in the literature.
David's Bloodhound tool processes lots of GBIF data for occurrences with names of those who collected or identified specimens. If you have an ORCID (and if you are a researcher you really should) then you can "claim" your specimens simply by logging in with your ORCID. My modest profile lists New Zealand crabs I collected while an undergraduate at Auckland University.

Unlike many biodiversity projects, Bloodhound is aimed squarely at individual researchers, it provides a means for you to show your contribution collecting and identifying the world's biodiversity. This raises the possibility of one day being able to add this information to your ORCID profile (in the way that currently ORCID can record your publications, data sets, and other work attached to a DOI). As David explains:
A significant contributing factor for this apparent neglect is the lack of a professional reward system; one that articulates and quantifies the breadth and depth of activities and expertise required to collect and identify specimens, maintain them, digitize their labels, mobilize the data, and enhance these data as errors and omissions are identified by stakeholders. If people throughout the full value-chain in natural history collections received professional credit for their efforts, ideally recognized by their administrators and funding bodies, they would prioritize traditionally unrewarded tasks and could convincingly self-advocate. Proper methods of attribution at both the individual and institutional level are essential.
Attribution at institutional level is an ongoing theme for natural history collections: how do they successfully demonstrate the value of their collections?
REMINDER: If you cite museum specimens in publications, let us know and send us a copy. WE LINK YOUR RESEARCH TO OUR COLLECTION FOREVER. It's like super important to science.
— Mark Carnall (@mark_carnall) October 24, 2018
Mark Carnall's (@mark_carnall) tweet illustrates the mismatch between a modern world of interconnected data and the reality of museums trying to track usage of their collections by requesting reprints. The idea of tracking citations of specimens and or collections has been around for a while. For example, I did some work text mining BioStor for museum specimen codes, Ross Mounce and Aime Rankin have worked on tracking citations of Natural History Museum specimens (https://github.com/rossmounce/NHM-specimens), and there is the clever use of Google Scholar by Winker and Withrow (see The impact of museum collections: one collection ≈ one Nobel Prize and https://doi.org/10.1038/493480b).
David has developed a nice tool that shows citations of specimens and/or collections from the Canadian Museum of Nature.

I'm sure many natural history collections would love a tool like this!
Note the altmetric.com "doughnuts" showing the attention each publication is receiving. These doughnuts are possible only because the publishing industry got together and adopted the same identifier system (DOIs). The existence of persistent identifiers enables a whole ecosystem to emerge based around those identifiers (and services to support those identifiers).
The biodiversity community has failed to achieve something similar, despite several attempts. Part of the problem is the cargo-cult obsession with "identifiers" rather than focussing on the bigger picture. So we have various attempts to create identifiers for specimens (see "Use of globally unique identifiers (GUIDs) to link herbarium specimen records to physical specimens" https://doi.org/10.1002/aps3.1027 for a review), but little thought given to how to build an ecosystem around those identifiers. We seem doomed to recreate all the painful steps publishers went through as created a menagerie of identifiers (e.g., SICIs, PII) and alternative linking strategies ("just in time" versus "just in case") until they settled on managed identifiers (DOIs) with centralised discovery tools (provided by CrossRef).
Specimen-level identifiers are potentially very useful, especially for cross linking records in GBIF, GenBank, and BOLD, as well as tracking citations, but not every taxonomic community has a history of citing specimens individually. Hence we may also want count citations at collection and institutional level. Once again we run into the issue that we lack persistent, widely used identifiers. The GRBio project to assign such identifiers has died, despite appeals to the community for support (see GRBio: A Call for Community Curation - what community?). Given Wikidata's growing role as an identity broker, a sensible strategy might be to focus on having every collection and institution in Wikidata (many are already) and add the relevant identifiers there. For example, Index Herbarium codes are now a recognised property in Wikidata, as seen in the entry for Cambridge University Herbarium (CGE).
But we will need more than technical solutions, we will also need compelling drivers to track specimen and collection use. The success of CrossRef has been due in part to the network effects inherent in the citation graph. Each publisher has a vested interest in using DOIs because other CrossRef members will include those DOIs in the list of literature cited, which means that each publisher potentially gets traffic from other members. Companies like altmetric.com (of doughnut fame) make money by selling data on attention papers receive to publishers and academic institutions, based on tracking mention of identifiers. Perhaps natural history collections should follow their lead and ask how they can get an equivalent system, in other words, how do we scale tools such as the Canadian Museum of Nature citation tracker across the whole network? And in particular, what services do you want and how much would those services be worth to you?
Ottawa Ecobiomics hackathon: graph databases and Wikidata
 I spent last week in Ottawa at a "Ecobiomics" hackathon organised by Joel Sachs. Essentially we spent a week exploring the application of linked data to various topics in biodiversity, with an emphasis on looking at working examples. Topics covered included:
I spent last week in Ottawa at a "Ecobiomics" hackathon organised by Joel Sachs. Essentially we spent a week exploring the application of linked data to various topics in biodiversity, with an emphasis on looking at working examples. Topics covered included:
- a live demo of Metaphacts
- a Semantic Mediawiki version of the Flora of North America
- using linked data to study food webs
- OpenBioDiv
In addition to the above I spent some of the time working on encoding GBIF specimen data in RDF with a view to adding this to Ozymandias. Having Steve Baskauf (@baskaufs) at the workshop was a great incentive to work on this, given his work with Cam Webb on Darwin-SW: Darwin Core-based terms for expressing biodiversity data as RDF.
A report is being written up which will discuss what we got up to in more detail, but one take away for me is the large cognitive burden that still stands in the way of widespread adoption of linked data approaches in biodiversity. Products such as Metaphactory go some way to hiding the complexity, but the overhead of linked data is high, and the benefits are perhaps less than obvious. Update: for more o this see Dan Brickley's comments on "Semantic Web Interest Group now closed".
In this context, the rise of Wikidata is perhaps the most important development. One thing we'd hoped to do but didn't get that far was to set up our own instance of Wikibase to play with (Wikibase is the software that Wikidata runs on). This is actually pretty straightforward to do if you have Docker installed, see this great post in Medium Wikibase for Research Infrastructure — Part 1 by Matt Miller, which I stumbled across after discovering Bob DuCharme's blog post Running and querying my own Wikibase instance. Running Wikibase on your own machine (if you follow the instructions you also get the SPARQL query interface) means that you can play around with a knowledge graph without worrying about messing up Wikidata itself, or having to negotiate with the Wikidata community if you want to add new properties. It looks like a relatively painless way to discover whether knowledge graphs are appropriate for the problem you're trying to solve. I hope to find time to play with Wikibase further in the future.
I'll update this blog post as the hackathon report is written.
GBIF Ebbe Nielsen Challenge update
🎉 🎉 CONGRATULATIONS to @UofGlasgow's @rdmpage for winning joint first prize in the 2018 @GBIF Ebbe Nielsen Challenge, the annual innovation competition to advance open science and open data for biodiversity. Well done, Prof Page! @IBAHCM #UofGWorldChangers pic.twitter.com/DjbmP90hTN
— UofG MVLS (@UofGMVLS) October 17, 2018
Quick note to express my delight and surprise that my entry for the 2018 GBIF Ebbe Nielsen Challenge come in joint first! My entry was Ozymandias - a biodiversity knowledge graph which built upon data from sources such as ALA, AFD, BioStor, CrossRef, ORCID), Wikispecies, and BLR.
I'm still tweaking Ozymandias, for example adding data on GBIF specimens (and maybe sequences from GenBank and BOLD) so that I can explore questions such as what is the lag time between specimen collection and description of a species. The bigger question I'm interested in is the extent to which knowledge graphs (aka RDF) can be used to explore biodiversity data.
For details on the other entries visit the list of winners at GBIF. The other first place winners Lien Reyserhove, Damiano Oldoni and Peter Desmet have generously donated half their prize to NumFOCUS which supports open source data science software:
Let’s give back! ❤️We decided to donate half of our @GBIF prize money (5000€) to @NumFOCUS which funds essential #opensource research software like @rOpenSci, #pandas and @ProjectJupyter. https://t.co/R3Bh0Tj8YX
— LifeWatch INBO (@LifeWatchINBO) October 18, 2018
This is a great way of acknowledging the debt many of us owe to developers of open source software that underpins the work of many researchers.
I hope GBIF and the wiser GBIF community found this year's Challenge to be worthwhile, I'm a big fan of anything which increases GBIF's engagement with developers and data analysts, and if the challenge runs again next year I encourage anyone with an interest in biodiversity informatics to consider taking part.
Tuesday, September 11, 2018
Guest post - Quality paralysis: a biodiversity data disease

In 2005, GBIF released Arthur Chapman's Principles of Data Quality and Principles and Methods of Data Cleaning: Primary Species and Species-Occurrence Data as freely available electronic publications. Their impact on museums and herbaria has been minimal. The quality of digitised collection data worldwide, to judge from the samples I've audited (see disclaimer below), varies in 2018 from mostly OK to pretty awful. Data issues include:
- duplicate records
- records with data items in the wrong fields
- records with data items inappropriate for a given field (includes Chapman's "domain schizophrenia")
- records with truncated data items
- records with items in one field disagreeing with items in another
- character encoding errors and mojibake
- wildly erroneous dates and spatial coordinates
- internally inconsistent formatting of dates, names and other data items (e.g. 48 variations on "sea level" in a single set of records)
In a previous guest post I listed 10 explanations for the persistence of messy data. I'd gathered the explanations from curators, collection managers and programmers involved with biodiversity data projects. I missed out some key reasons for poor data quality, which I'll outline in this post. For inspiration I'm grateful to Rod Page and to participants in lively discussions about data quality at the SPNHC/TDWG conference in Dunedin this August.
- Our institution, like all natural history collections these days, isn't getting the curatorial funding it used to get, but our staff's workload keeps going up. Institution staff are flat out just keeping their museums and herbaria running on the rails. Staff might like to upgrade data quality, but as one curator wrote to me recently, "I simply don't have the resources necessary."
- We've been funded to get our collections digitised and/or online, but there's nothing in the budget for upgrading data quality. The first priority is to get the data out there. It would be nice to get follow-up funding for data cleaning, but staff aren't hopeful. The digitisation funder doesn't seem to think it's important, or thinks that staff can deal with data quality issues later, when the digitisation is done.
- There's no such thing as a Curator of Data at our institution. Collection curators and managers are busy adding records to the collection database, and IT personnel are busy with database mechanics. The missing link is someone on staff who manages database content. The bigger the database, the greater the need for a data curator, but the usual institutional response is "Get the collections people and the IT people together. They'll work something out."
- Aggregators act too much like neutrals. We're mobilising our data through an aggregator, but there are no penalties if we upload poor-quality data, and no rewards if we upload high-quality data. Our aggregator has a limited set of quality tests on selected data fields and adds flags to individual records that have certain kinds of problems. The flags seem to be mainly designed for users of our data. We don't have the (time/personnel/skills) to act on this "feedback" (or to read those 2005 GBIF reports).
There's a 15th explanation that overlaps the other 14 and Rod Page has expressed it very clearly: there's simply no incentive for anyone to clean data.
- Museums and herbaria don't get rewards, kudos, more visitors, more funding or more publicity if staff improve the quality of their collection data, and they don't get punishments, opprobrium, fewer visitors, reduced funding or less publicity if the data remain messy.
- Aggregators likewise. Aggregators also don't suffer when they downgrade the quality of the data they're provided with.
- Users might in future get some reputational benefit from alerting museums and herbaria to data problems, through an "annotation system" being considered by TDWG. However, if users clean datasets for their own use, they get no reward for passing blocks of cleaned data to overworked museum and herbarium staff, or to aggregators, or to the public through "alternative" published data versions.
With the 15 explanations in mind, we can confidently expect collection data quality to remain "mostly OK to pretty awful" for the foreseeable future. Data may be upgraded incrementally as loans go out and come back in, and as curators, collection managers and researchers compare physical holdings one-by-one with their digital representations. Unfortunately, the improvements are likely to be overwhelmed by the addition of new, low-quality records. Very few collection databases have adequate validation-on-entry filters, and staff don't have time for, or assistance with checking. Or a good enough reason to check.
"Quality paralysis" is endemic in museums and herbaria and seems likely to be with us for a long time to come.
DISCLAIMER: Believe it or not, this post isn't an advertisement for my data auditing services.
I began auditing collection data in 2012 for my own purposes and over the next few years I offered free data auditing to a number of institutions in Australia and elsewhere. There were no takers.
In 2017 I entered into a commercial arrangement with Pensoft Publishers to audit the datasets associated with data papers in Pensoft journals, as a free Pensoft service to authors. Some of these datasets are based on collections data, but when auditing I don't deal with the originating institutions directly.
I continue to audit publicly available museum and herbarium data in search of raw material for my website A Data Cleaner's Cookbook and its companion blog BASHing data. I also offer free training in data auditing and cleaning.
Monday, August 20, 2018
GBIF Challenge Entry: Ozymandias
I've submitted an entry for the 2018 GBIF Ebbe Nielsen Challenge. It's a couple of weeks before the deadline but I will be away then so have decided to submit early.
My entry is Ozymandias - a biodiversity knowledge graph. The name is a play on "Oz" being nickname for Australia (much of the data for the entry comes from Australia), and Ozymandias, which is a poem about hubris, and attempting to link biodiversity data requires a certain degree of hubris.
The submission process for the challenge is unfortunately rather opaque compared to previous years when entries were visible to all, so participants could see what other people were submitting, and also knew the identity of the judges, etc. In the spirit of openness here is my video summarising my entry:
Ozymandias - GBIF Challenge Entry from Roderic Page on Vimeo.
There is also a background document here: https://docs.google.com/presentation/d/1UglxaL-yjXsvgwn06AdBbnq-HaT7mO4H5WXejzsb9MY/edit?usp=sharing.I suspect this entry is not at all what the challenge is looking for, but I've used the challenge as a deadline so that I get something out the door rather than endlessly tweaking a project that only I can see. There will, of course, be endless tweaking as I explore further ways to link data, but at least this way there is something people can look at. Now, I need to spend some time writing up the project, which will require yet more self discipline to avoid the endless tweaking.
Friday, August 17, 2018
Ozymandias demo
I've made a video walkthrough of Ozymandias, which I described in this post. It's a bit, um, long, so I'll need to come up with a shorter version.
Ozymandias - a biodiversity knowledge graph from Roderic Page on Vimeo.
Friday, August 10, 2018
Ozymandias: a biodiversity knowledge graph of Australian taxa and taxonomic publications
In the spirit of release early and release often, here is the first workable version of a biodiversity knowledge graph that I've been working on for Australian animals (for some background on knowledge graphs see Towards a biodiversity knowledge graph now in RIO). The core of this knowledge graph is a classification of animals from the Atlas of Living Australia (ALA) combined with data on taxonomic names and publications from the Australian Faunal Directory (AFD). This has been enhanced by adding lots of digital identifiers (such as DOIs) to the publications and, where possible, full text either as PDFs or as page scans from the Biodiversity Heritage Library (BHL) (provided via BioStor). Identifiers enable us to further grow the knowledge graph, for example by adding "cites" and "cited by" links between publications (data from CrossRef), and displaying figures from the Biodiversity Literature Repository (BLR).
The demo is here: https://ozymandias-demo.herokuapp.com/ If you’re looking for starting points, you could try:
Assassin spiders (images from Plazi and citation data from CrossRef) https://ozymandias-demo.herokuapp.com/?uri=https://biodiversity.org.au/afd/publication/64908f75-456b-4da8-a82b-c569b4806c22

Memoirs of Museum Victoria (dynamic query finds record in Wikidata and adds map) https://ozymandias-demo.herokuapp.com/?uri=https://biodiversity.org.au/afd/publication/5c22a8d1-7456-4f8c-9384-1246ecbf15a6

G. R. Allen (we can from the taxonomic tree of his top 20 taxa that he studies fish - who knew?) https://ozymandias-demo.herokuapp.com/?uri=https://biodiversity.org.au/afd/publication/%23creator/g-r-allen

Paper on mosquito taxonomy with lots of citations, including material in BHL/BioStor https://ozymandias-demo.herokuapp.com/?uri=https://biodiversity.org.au/afd/publication/578d1dec-5816-49ec-8916-3f957fd230f5

Paper on Australian flies with full text in BioStor https://ozymandias-demo.herokuapp.com/?uri=https://biodiversity.org.au/afd/publication/0ffe4f28-b8ac-4132-be34-19eb03fbf685

The focus for now is on taxa, publications, journals, and people. Occurrences and sequences are on the “to do” list. As always there’s lots of data cleaning and cross linking to do, but an obvious next step is to link people’s names to identifiers such as ORCID and Wikidata ids, so that we can trace the activities of taxonomists as they discover and describe Australian biodiversity (the choice of Australia is simply to keep things manageable, and because the amount of data and digitisation they’ve done is pretty extraordinary). I’m also working to a deadline as I'm trying to get this demo wrapped up in the next couple of weeks.
Technical details
TL;DR the knowledge graph is implemented as a triple store where the data has been represented using a small number of vocabularies (mostly schema.org with some terms borrowed from TAXREF-LD and the TDWG LSID vocabularies). All results displayed in the first two panels are the result of SPARQL queries, the content in the rightmost panel comes from calls to external APIs. Search is implemented using Elasticsearch. If you are feeling brave you can query the knowledge graph directly in SPARQL. I’m constantly tweaking things and adding data and identifiers, so things are likely to break. More details and documentation will be going up on the GitHub repository.Friday, July 20, 2018
Signals from Singapore: NGS barcoding, generous interfaces, the return of faunas, and taxonomic burden
 Earlier this year I stopped over in Singapore, home of the spectacular "supertrees" in the Garden by the Bay. The trip was a holiday, but I spent a good part of one day visiting Rudolf Meier's group at the National University of Singapore. Chatting with Rudolf was great fun, he's opinionated and not afraid to share those opinions with anyone who will listen. Belatedly I've finally written up some of the topics we discussed.
Earlier this year I stopped over in Singapore, home of the spectacular "supertrees" in the Garden by the Bay. The trip was a holiday, but I spent a good part of one day visiting Rudolf Meier's group at the National University of Singapore. Chatting with Rudolf was great fun, he's opinionated and not afraid to share those opinions with anyone who will listen. Belatedly I've finally written up some of the topics we discussed.
Massively scalable and cheap DNA barcoding
Singapore has a rich fauna in a small area, full of undescribed species, so DNA barcoding seems an obvious way to get a handle on its biodiversity. Rudolf has been working towards scalable and cheap barcoding, e.g. $1 DNA barcodes for reconstructing complex phenomes and finding rare species in specimen‐rich samples https://doi.org/10.1111/cla.12115 . His lab can sequence short (~300 bp) barcode sequences for around $US 0.50 per specimen. Their pipeline generates lots of data, accompanied by high quality photographs of exemplar specimens, which contribute to The Biodiversity of Singapore, a "Digital Reference Collection for Singapore's Biodiversity". This site provides a simple but visually striking way to explore Singapore's biota, and is a nice example of what Mitchell Whitelaw calls "generous interfaces". We could do with more of these for biodiversity data.
One nice feature of regular COI DNA barcodes is that they are comparable across labs because everyone is sequencing the same stretch of DNA. With short barcodes, different groups may target different regions of the COI gene, resulting in sequences that can't be compared. For example, the 127bp mini barcodes developed in A universal DNA mini-barcode for biodiversity analysis https://doi.org/10.1186/1471-2164-9-214 are completely disjoint from the ~300bp sequenced by Meier's group (I'm trying to keep track of some of these short barcodes here: https://gist.github.com/rdmpage/4f2545eeea4756565925fb4307d9af6b.
The return of regional faunas
In the "old days" of colonial expansion it was common for taxonomists to write volume entitled "The Fauna of [insert colonised country here]". These were regional works focussing on a particular area, often motivated by trying to catalogue animals of potential economic or medical importance, as well as of scientific interest. By limiting their geographic scope, faunal treatments of taxa can sometimes be inadequate. Descriptions of new species from a particular area may be hard to compare with descriptions of species in the same group that occur elsewhere and are described by other taxonomists. It may be that to do the taxonomy of a particular group well you need to treat that group throughout its geographic range, rather then just those species in your geographic area. Hence faunas loose their scientific appeal, despite the attractiveness of having a detailed summary of the fauna of a particular area. DNA sequencing circumvents this problem by having a universally comparable character. You can sequence everything within a geographic region, but those sequences will be directly comparable to sequences found elsewhere. Barcoding makes faunas attractive again, which may help funding taxonomic research because it makes funding projects with a restricted national scope scientifically still worthwhile.Taxonomic burden and legacy names
As we discover and catalogue more and more of the planet's biodiversity we want to stick names on that biodiversity, and this can be a significant challenge when there is a taxonomic legacy of names that are so poorly described it is hard to establish how they relate to the material we are working with. Even if you have access to the primary literature through digitisation projects like BHL, if the descriptions are poor, if the types are lost or their identity is confused (see for example A New Species of Megaselia Rondani (Diptera: Phoridae) from the Bioscan Project in Los Angeles, California, with Clarification of Confused Type Series for Two Other Species https://doi.org/10.4289/0013-8797.118.1.93 by Emily A. Hartop - who I met on this trip - and colleagues), or can't be sequenced, then these names will remain ambiguous, and potentially clogging up efforts to name the unnamed species. One approach favoured by Rudolf is to effectively wipe the slate clean, declare all ambiguous names before a certain date to be null and void, and start again. This renders (or rather, resets) the notion of priority - given two names for the same species the older name is the one to use - and so is likely to be a hard sell, but it is part of the ongoing discussion about the impact of molecular data on naming taxa. Similar discussions are raging at the moment in mycology, e.g. Ten reasons why a sequence-based nomenclature is not useful for fungi anytime soon https://doi.org/10.5598/imafungus.2018.09.01.11, yet a another reflection of how much taxonomy is driven by technology.Thursday, July 05, 2018
GBIF at 1 billion - what's next?
GBIF has reached 1 billion occurrences which is, of course, something to celebrate:
#GBIF1billion has arrived! Merci beaucoup, @Le_Museum @INPN_MNHN et @gbiffrance!
— GBIF (@GBIF) July 4, 2018
Thanks and congratulations, too, to the 1,217 data publishers and 92 participants who make the GBIF network go! More details to follow Thursday (champagne doesn't drink itself)… pic.twitter.com/xQ2f5fIt2x
An achievement on this scale represents a lot of work by many people over many years, years spent developing simple standards for sharing data, agreeing that sharing is a good thing in the first place, tools to enable sharing, and a place to aggregate all that shared data (GBIF).
So, I asked a question:
So I guess the real #GBIF1billion question is what can we do with a billion data points that we couldn't do with, say, a hundred million? Does more data simply mean more of same kind of analyses, or does it enable something new (and exciting)? @GBIF
— Roderic Page (@rdmpage) July 4, 2018
My point is not to do this:
Hey, don't spoil the party!
— Dimitri Brosens (@Dimibro) July 4, 2018
Rather it is to encourage a discussion about what happens when we have large amounts of biodiversity data. Is it the case that as we add data we simply enable more of the same kind of science, only better (e.g., more data for species distribution modelling), or do we reach a point where new things become possible?
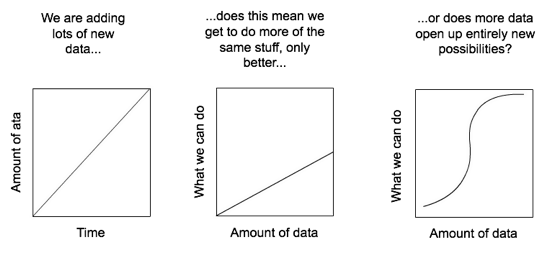
To give a concrete example, consider iNaturalist. This started out as a Masters project to collect photos of organisms on Flickr. As you add more images you get better coverage of biodiversity, but you still have essentially a bunch of pictures. But once you have LOTS of pictures, and those are labelled with species names, you reach the point where it is possible to do something much more exciting - automatic species identification. To illustrate, I recently took the photos below:


Note the reddish tubular growths on the leaves. I asked iNaturalist to identify these photos and within a few seconds it came back with Eriophyes tiliae, the Red Nail Gall Mite. This feels like magic. It doesn't rely on complicated analysis of the image (as many earlier efforts at automated identification have done) it simply "knows" that images that look like this are typically of the galls of this mite because it has seen many such images before. (Another example of the impact of big data is Google Translate, initially based on parsing lots of examples of the same text in multiple languages.)
Okay, but then not sure I see what you're looking for. Why would 1 billion, as opposed to, say, 100 million, mean a paradigm shift? Do you have any (even hypothetical) answers to suggest yourself?
— Leif Schulman (@Leif_Sch) July 5, 2018
The "1 billion" number is not, by itself, meaningful. It's rather that I hope that while we're popping the champagne and celebrating a welcome, if somewhat arbitrary milestone, I'm hoping that someone, somewhere is thinking about whether biodiversity data on this scale enables something new.
Do I have answers? Not really, but here's one fairly small-scale example. One of the big challenges facing GBIF is getting georeferenced data. We spend a lot of time using a variety of tools and databases to convert text descriptions one collection localities into latitude and longitude. Many of these descriptions include phrases such as "5 mi NW of" and so we've developed parsers to attempt to make sense of these. All of these phrases and the corresponding latitude and longitude coordinates have ended up in GBIF. Now, this raises the possibility that after a point, pretty much any locality phrase will be in GBIF, so a way to georeference a locality is simply to search GBIF for that locality and use the associated latitude and longitude. GBIF itself becomes the single best tool to georeference specimen data. To explore this idea I've built a simple tool on glitch https://lyrical-money.glitch.me that takes a locality description and geocodes it using GBIF.

You paste in a locality string and it attempt to find that on a map based on data in GBIF. This could be automated, so you could imagine being able to georeference whole collections as part of the process of uploading the data to GBIF. Yes, the devil is in the details, and we'd need ways to flag errors or doubtful records, but the scale of GBIF starts of open up possibilities like this.
So, my question is, "what's next?".
Wednesday, June 13, 2018
Liberating links between datasets using lightweight data publishing: an example using IPNI and the taxonomic literature
 I've written a short paper entitled "Liberating links between datasets using lightweight data publishing: an example using plant names and the taxonomic literature" (phew) and put a preprint on bioRxiv (https://doi.org/10.1101/343996) while I figure out where to publish it. Here's the abstract:
I've written a short paper entitled "Liberating links between datasets using lightweight data publishing: an example using plant names and the taxonomic literature" (phew) and put a preprint on bioRxiv (https://doi.org/10.1101/343996) while I figure out where to publish it. Here's the abstract:
Constructing a biodiversity knowledge graph will require making millions of cross links between diversity entities in different datasets. Researchers trying to bootstrap the growth of the biodiversity knowledge graph by constructing databases of links between these entities lack obvious ways to publish these sets of links. One appealing and lightweight approach is to create a "datasette", a database that is wrapped together with a simple web server that enables users to query the data. Datasettes can be packaged into Docker containers and hosted online with minimal effort. This approach is illustrated using a dataset of links between globally unique identifiers for plant taxonomic names, and identifiers for the taxonomic articles that published those names.
In some ways the paper is simply a record of me trying to figure out how to publish a project that I've been working on for several years, namely linking names from BioNames. The preprint discusses various options, before settling on "datasettes", which is a nice method developed by Simon Willison (@simonw) to wrap up simple databases with their own web server and query API and make them accessible on the web. These can run on a local machine, or be packaged up as a Docker container, which is what I've done. You play with the database here: https://ipni.sloppy.zone. If this link is offline, then you can grab the container here https://hub.docker.com/r/rdmpage/ipni/ and run it yourself. If, like me, you're new to Docker, then I recommend grabbing a copy of Kitematic.
The datasette interface is simple but gives you lots of freedom to explore the data.

For example, you have ability to query the data using SQL, e.g.:
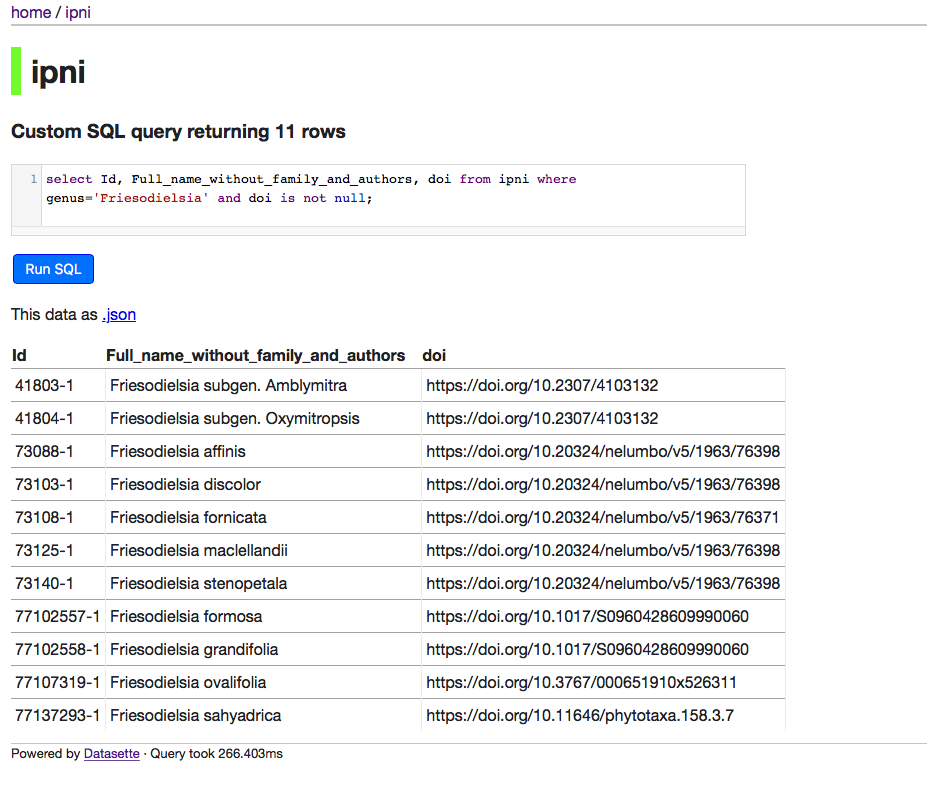
One advantage of this approach is that the data is more accessible. I could just dump the database somewhere but then you'd have to download a large file and figure out how query it. This way, you can play with it straight away. It also means people can make use of it before I make up my mind how best to package it (for example, as part of a larger database of eukaryote names). This is one of the main motivations behind the paper, how to avoid the trap of spending years cleaning and augmenting data and not making it available to others because of the overhead of building a web site around the data. I may look at liberating some other datasets using this approach.
Monday, June 04, 2018
Towards a biodiversity token: Bitcoin, FinTech, and a radical suggestion for the GBIF Challenge
 First off, let me say that what follows is a lot of arm waving to try and obscure how little I understand what I'm talking about. I'm going to sketch out what I think is a "radical" idea for a GBIF Challenge entry.
First off, let me say that what follows is a lot of arm waving to try and obscure how little I understand what I'm talking about. I'm going to sketch out what I think is a "radical" idea for a GBIF Challenge entry.
TL;DR GBIF should issue it's own cryptocurrency and use that to fund the development of the GBIF network by charging for downloading cleaned, processed data (original provider data remains free). People can buy subscriptions to get access to data, and/or purchase GBIF currency as a contribution or investment. Proceeds from the purchase of cleaned data are divided between GBIF (to fund the portal), the data providers (to reward them making data available) and the GBIF nodes in countries included in the geographic coverage of the data (to help them build their biodiversity infrastructure). The challenge entry would involve modelling this idea and conducting simulations to test it's efficacy.
The motivation for this idea comes from several sources:
1. GBIF is (under-)funded by direct contributions from governments, hence each year it essentially "begs" for money. Several rich countries (such as the United Kingdom) struggle to pay the fairly paltry sums involved. Part of the problem is that they are taking something of demonstrable value (money) and giving it to an organisation (GBIF) which has no demonstrable financial value. Hence the argument for funding GBIF is basically "it's the right thing to do". This is not really a tenable or sustainable model.
2. Many web sites provide information for "free" in that the visitor doesn't pay any money. Instead the visitor views ads and, whether they are aware if it or not, are handing over large amounts of data about themselves and their behaviour (think the recent scandal involving Facebook).
3. Some people are rebelling against the "free with ads" by seeking other ways to fund the web. For example, the Brave web browser enables you to buy BATS (Basic Attention Tokens, based on Ethereum). You can choose to send BATS to web sites that you visit (and hence find valuable). Those sites don't need to harvest tyour data or bombard you with ads to receive an income.
4. Cryptocurrency is being widely explored as a way to raise funding for new ventures. Many of these are tech-based, but there are some interesting developments in conservation and climate change, such as Veridium which offsets carbon emissions. There are links between efforts like Veridium and carbon offset programmes such as the Rimba Raya Biodiversity Reserve, so you can go from cryptocurrency to trees.
5. The rather ugly, somewhat patronising furore that erupted when Rwanda decided that the best way to increase its foreign currency earnings (as a step towards ultimately freeing itself from dependency on development aid) was to sign a sponsorship deal with Arsenal football club.
Now, imagine a situation where GBIF has a cryptocurrency token (e.g., the "GBIF coin"). Anyone, whether a country, an organisation, or an individual can buy GBIF coins. If you want to download GBIF data, you will need to pay in GBIF coins, either per-download or via a monthly subscription. The proceeds from each download are split in a way that supports the GBIF network as a whole. For example, imagine GBIF itself gets 30% (like Apple's App Store). The remaining 70% gets' split between (a) the data providers and (b) the GBIF nodes in countries included in the data download. For example, almost all the data on a country such as Rwanda does not come from Rwanda itself, but from other countries. You want to reward anyone who makes data available, but you also want to support the development of a biodiversity data infrastructure in Rwanda (or any other country), so part of the proceeds go to the GBIF node in Rwanda.
Now, an immediate issue (apart from the merits or otherwise of blockchains and cryptocurrency) is that I'm advocating charging for access to data, which seems antithetical to open access. To be clear, I think open access is crucial. I'm suggesting that we distinguish between two classes of data. The first is the data as it is provided to GBIF. That is almost always open data under a CC0 license, and that remains free. But if you ant it for free it is served as it is received. In other words, for free access to data GBIF is essentially a dumb repository (like, say, Dryad). The data is there, you can search the metadata for each dataset, so essentially you get something like the current dataset search.
The other thing GBIF does is that it processes the data, cleaning it, reconciling names and locations, and indexing it, so that if you want to search for a given species, GBIF summarises the data across all the datasets and (often) presents you with a better result that if you'd downloaded all the original data and simply merged it together yourself. This is a valuable service, and its one of the reasons why GBIF costs money to run. So imagine that we do something like this:
- It is free to browse GBIF as a person and explore the data
- It is free to download the raw data provided by any data publisher.
- It costs to download cleaned data that corresponds to a specific query, e.g. all records for a particular taxon, geographic area, etc.
- Payment for access to cleaned data is via the GBIF coin.
- The cost is small, on the scale of buying a music track or subscribing to Spotify.
Now, I don't expect GBIF to embrace this idea anytime soon. By nature it's a conservative, risk-averse organisation. But I think something like this idea deserves serious attention, ideally from people with much better understanding of the issues that my own "I saw this on Twitter therefore it must be cool" level. One way to move forward would be to model how such a system would work, based for example on data on web site visits and data downloads on the current GBIF portal. I suspect models could be built to give some idea of whether such an approach would be financially viable. It occurs to me that something like this would make a great GBIF Challenge entry, particularly as it is gives a license for thinking the unthinkable with no risk to GBIF itself.
Wednesday, May 09, 2018
World Taxonomists and Systematists via ORCID
 David Shorthouse (@dpsspiders) makes some very cool things, and his latest project World Taxonomists & Systematists is a great example of using automation to assemble a list of the world's taxonomists and systematists. The project uses ORCID. As many researchers will know, ORCID's goal is to have every researcher uniquely identified by an ORCID id (mine is https://orcid.org/0000-0002-7101-9767) that is linked to all a researcher's academic output, including papers, datasets, and more. So David has been querying ORCID for keywords such as taxonomist, taxonomy, nomenclature, or systematics to locate taxonomists and add them to his list. For more detail see his post on the ORCID blog.
David Shorthouse (@dpsspiders) makes some very cool things, and his latest project World Taxonomists & Systematists is a great example of using automation to assemble a list of the world's taxonomists and systematists. The project uses ORCID. As many researchers will know, ORCID's goal is to have every researcher uniquely identified by an ORCID id (mine is https://orcid.org/0000-0002-7101-9767) that is linked to all a researcher's academic output, including papers, datasets, and more. So David has been querying ORCID for keywords such as taxonomist, taxonomy, nomenclature, or systematics to locate taxonomists and add them to his list. For more detail see his post on the ORCID blog.
Using ORCIDs to help taxonomists gain visibility is an idea that's been a round for a little while. I blogged about it in Possible project: #itaxonomist, combining taxonomic names, DOIs, and ORCID to measure taxonomic impact, at which time David was already doing another cool piece of work linking collectors to ORCIDs and their collecting effort, see e.g. data for Terry A. Wheeler.
There are, of course, a bunch of obstacles to this approach. Many taxonomists lack ORCIDs, and I keep coming across "private" ORCIDs where taxonomists have an ORCID id but don't make their profile public, which makes it hard to identify them as taxonomists. Typically I discover these profiles via metadata in CrossRef, which will list the ORCID id for any authors that have them and have made them know to the publisher of their paper.
ORCID ids are only available for people who are alive (or alive recently enough to have registered), so there will be many taxonomists who will never have an ORCID id. In this case, it may be Wikidata to the rescue:
Here for example is a SPARQL query for all taxon authors with no date of deathhttps://t.co/xlgLG0Md3m
— Andy Mabbett (@pigsonthewing) April 6, 2018
(Just hit the big "arrow" button to run it - takes a short while; 16K results)
H/T @WikidataFacts for the query.
Many taxonomists have Wikidata entries because they are either notable enough to be in Wikipedia, or they have an entry in Wikispecies, and people like Andy Mabbett (@pigsonthewing) have been diligently ensuring these people have Wikidata entries. There's huge scope for making use of these links.
Meanwhile, if you are a taxonomist or a systematist and you don't have an ORCID, get yourself one at ORCID, claim your papers, and you should appear shortly in the World Taxonomists & Systematists list.
2018 GBIF Ebbe Nielsen Challenge now open
 Last year I finished my four-year stint as Chair of the GBIF Science Committee. During that time, partly as a result of my urging, GBIF launched an annual "GBIF Ebbe Nielsen Challenge", and I'm please that this year GBIF is continuing to run the challenge. In 2015 and 2016 the challenge received some great entries.
Last year I finished my four-year stint as Chair of the GBIF Science Committee. During that time, partly as a result of my urging, GBIF launched an annual "GBIF Ebbe Nielsen Challenge", and I'm please that this year GBIF is continuing to run the challenge. In 2015 and 2016 the challenge received some great entries.
Last year's challenge (GBIF Challenge 2017: Liberating species records from open data repositories for scientific discovery and reuse didn't attract quite the same degree of attention, and GBIF quietly didn't make an award. I think part of the problem was that there's a fine balance between having a wide open challenge which attracts all sorts of interesting entries, some a little off the wall (my favourite was GBIF data converted to 3D plastic prints for physical data visualisation) versus a specific topic which might yield one or more tools that could, say, be integrated into the GBIF portal. But if you make it too narrow then you run the risk of getting fewer entries, which is what happened in 2017. Ironically, since the 2017 challenge I've come across work that would have made a great entry, such as a thesis by Ivelize Rocha Bernardo Promoting Interoperability of Biodiversity Spreadsheets via. Purpose Recognition, see also Bernardo, I. R., Borges, M., Baranauskas, M. C. C., & Santanchè, A. (2015). Interpretation of Construction Patterns for Biodiversity Spreadsheets. Lecture Notes in Business Information Processing, 397–414. doi:10.1007/978-3-319-22348-3_22.
This year the topic is pretty open:
The 2018 Challenge will award €34,000 for advancements in open science that feature tools and techniques that improve the access, utility or quality of GBIF-mediated data. Under this open-ended call, challenge submissions may build on existing tools and features, such as the GBIF API, Integrated Publishing Toolkit, data validator, relative species occurrence tool, among others—or develop new applications, methods, workflows or analyses.
Lots of scope, and since I'm not longer part of the GBIF Science Committee it's tempting to think about taking part. The judging criteria are pretty tough and results-oriented:
Winning entries will demonstrably extend and increase the usefulness, openness and visibility of GBIF-mediated data for identified stakeholder groups. Each submission is expected to demonstrate advantages for at least three of the following groups: researchers, policymakers, educators, students and citizen scientists.
So, maybe less scope for off-the-wall stuff, but an incentive to clearly articulate why a submission matters.
The actual submission process is, sadly, rather more opaque than in previous years where it was run in the open on Devpost where you can still see previous submissions (e.g., those for 2015). Devpost has lots of great features but isn't cheap, so the decision is understandable. Maybe some participants will keep the rest of the community informed via, say, Twitter, or perhaps people will keep things close to their chest. In any event, I hope the 2018 challenge inspires people to think about doing something both cool and useful with biodiversity data. Oh, and did I mention that a total of €34,000 in prizes is up for grabs? Deadline for submission is 5 September 2018.
iSpecies meets Lifemap
It's been a little quiet on this blog as I've been teaching, and spending a lot of time data wrangling and trying to get my head around "data lakes" and "triple stores". So there are a few things to catch up on, and a few side projects to report on.
I continue to play with iSpecies, which is a simple mashup off biodiversity data sources. When I last blogged about iSpecies I'd added TreeBASE as a source (iSpecies meets TreeBASE). iSpecies also queries Open Tree of Life, and I've always wanted a better way of displaying the phylogenetic context of a species or genus. TreeBASE is great for a detailed, data-driven view, but doesn't put the taxon in a larger context, nor does the simple visualisation I developed for Open Tree of Life.
A nice large-scale tree visualisation is Lifemap (see De Vienne, D. M. (2016). Lifemap: Exploring the Entire Tree of Life. PLOS Biology, 14(12), e2001624. doi:10.1371/journal.pbio.2001624), and it dawned on me that since Lifemap uses the same toolkit (leaflet.js) that I use to display a map of GBIF records, I could easily add it to iSpecies. After looking at the Lifemap HTML I figured out the API call I need to pan the map to given taxon using Open Tree of Life taxon identifiers, and violà, I now have a global tree of life that shows where the query taxon fits in that tree.
Here's a screenshot of a search for Podocarpus showing the first 300 records from GBIF, and the position of Podocarpus in the tree of life. The tree is interactive so you can zoom and pan just like the GBIF map.

Here's another one for the genus Timonius:

Very much still at the "quick and dirty" stage, but I continue to marvel at how much information can be assembled "on the fly" from a few sources, and how much richer this seems than what biodiversity informatics projects offer. There's a huge amount of information that is simpy being missed or under-utilised in this area.
Wednesday, January 24, 2018
Guest post: The Not problem

Nico Franz and Beckett Sterner created a stir last year with a preprint in bioRxiv about expert validation (or the lack of it) in the "backbone" classifications used by aggregators. The final version of the paper was published this month in the OUP journal Database (doi:10.1093/database/bax100).
To see what effect "backbone" taxonomies are having on aggregated occurrence records, I've recently been auditing datasets from GBIF and the Atlas of Living Australia. The results are remarkable, and I'll be submitting a write-up of the audits for formal publication shortly. Here I'd like to share the fascinating case of the genus Not Chan, 2016.
I found this genus in GBIF. A Darwin Core record uploaded by the New Zealand Arthropod Collection (NZAC02015964) had the string "not identified on slide" in the scientificName field, and no other taxonomic information.
GBIF processed this record and matched it to the genus Not Chan, 2016, which is noted as "doubtful" and "incertae sedis".
There are 949 other records of this genus around the world, carefully mapped by GBIF. The occurrences come from NZAC and nine other datasets. The full scientific names and their numbers of GBIF records are:
| Number | Name |
|---|---|
| 2 | Not argostemma |
| 14 | not Buellia |
| 1 | not found, check spelling |
| 1 | Not given (see specimen note) bucculenta |
| 1 | Not given (see specimen note) ortoni |
| 1 | Not given (see specimen note) ptychophora |
| 1 | Not given (see specimen note) subpalliata |
| 1 | not identified on slide |
| 1 | not indentified |
| 1 | Not known not known |
| 1 | Not known sp. |
| 1 | not Lecania |
| 4 | Not listed |
| 873 | Not naturalised in SA sp. |
| 18 | Not payena |
| 5 | not Punctelia |
| 18 | not used |
| 6 | Not used capricornia Pleijel & Rouse, 2000 |
GBIF cites this article on barnacles as the source of the genus, although the name should really be Not Chan et al., 2016. A careful reading of this article left me baffled, since the authors nowhere use "not" as a scientific name.
Next I checked the Catalogue of Life. Did CoL list this genus, and did CoL attribute it to Chan? No, but "Not assigned" appears 479 times among the names of suprageneric taxa, and the December 2018 CoL checklist includes the infraspecies "Not diogenes rectmanus Lanchester,1902" as a synonym.
The Encyclopedia of Life also has "Not" pages, but these have in turn been aggregated on the "EOL pages that don't represent real taxa" page, and under the listing for the "Not assigned36" page someone has written:
This page contains a bunch of nodes from the EOL staff Scratchpad. NB someone should go in and clean up that classification.
"Someone should go in and clean up that classification" is also the GBIF approach to its "backbone" taxonomy, although they think of that as "we would like the biodiversity informatics community and expert taxonomists to point out where we've messed up". Franz and Sterner (2018) have also called for collaboration, but in the direction of allowing for multiple taxonomic schemes and differing identications in aggregated biodiversity data. Technically, that would be tricky. Maybe the challenge of setting up taxonomic concept graphs will attract brilliant developers to GBIF and other aggregators.
Meanwhile, Not Chan, 2016 will endure and aggregated biodiversity records will retain their vast assortment of invalid data items, character encoding failures, incorrect formatting, duplications and truncated data items. In a post last November on the GitHub CoL+ pages I wrote:
Being old and cynical, I can speculate that in the time spent arguing the "politics" of aggregation in recent years, a competent digital librarian or data scientist would have fixed all the CoL issues and would be halfway through GBIF's. But neither of those aggregators employ digital librarians or data scientists, and I'm guessing that CoL+ won't employ one, either.