The new look Biodiversity Heritage Library includes articles extracted from BioStor, which is a step forwards in making the "legacy" biodiversity literature more accessible. But we still have some way to go. In particular the articles lack the obvious decoration of a modern article, the DOI. Consequently these articles still live in a twilight zone where they are cited in the literature but not linked to. DOIs are becoming more common for taxonomic articles. Zookeys has them, and now Zootaxa has adopted them (and will be applying them retrospectively to thousands of already published articles). Major archives of back issues digitised by Taylor and Francis, and Wiley, for example, also have DOIs.
One obstacle to assigning CrossRef DOIs to articles in BHL is the convention that DOIs are typically managed by the publisher of the journal. But in a number of cases the publisher may no longer exist, the journal may no longer be published, or the publisher may lack the commercial resources to support DOIs. In these cases perhaps BHL could adopt the role of publisher?
Another approach is that adopted by a number of other digital archives, whereby the archive assigns DOIs to articles, but these DOIs are registered not through CrossRef but with another DOI registration agency, such as DataCite. For example the Swiss Electronic Academic Library Service (SEALS) archive assigns DOIs to individual articles, such as http://dx.doi.org/10.5169/seals-88913.
There are some limitations to not using CrossRef DOIs, in particular, you don't get the full benefits of their metadata-based services such as getting metadata from a DOI, discovering DOIs from metadata, or citation linking. But all is not lost. Some services support both CrossRef and DataCite DOIs, such as http://crosscite.org/citeproc. For example, for the DOI 10.5169/seals-88913 we get some basic formatting:
Perret, Jean-Luc. (1961). Etudes herpétologiques africaines III. Société Neuchâteloise des Sciences Naturelles. doi:10.5169/seals-88913
This still leaves us lacking some services, such as finding DOIs for articles cited in a manuscript. However this is a service we can provide, and will have to anyway if we want to find all the digitised literature available (e.g., archives such as SEALS as well as numerous instances of DSpace). My preference would be for CrossRef DOIs, but if that proves problematics we can still get much of the functionality we need using other DOI providers.
Rants, raves (and occasionally considered opinions) on phyloinformatics, taxonomy, and biodiversity informatics. For more ranty and less considered opinions, see my Twitter feed.
ISSN 2051-8188. Written content on this site is licensed under a Creative Commons Attribution 4.0 International license.
Tuesday, March 26, 2013
Towards DOIs for Biodiversity Heritage Library articles
Location:
Wengern Wengern
Wednesday, March 20, 2013
BioNames update - matching taxon names to classifications
On eof the things BioNames will need to do is match taxon names to classifications. For example, if I want to display a taxonomic hierarchy for the user to browse through the names, then I need a map between the taxon names that I've collected and one or more classifications. The approach I'm taking is to match strings, wherever possible using both the name and taxon authority. In many cases this is straightforward, especially if there is only one taxon with a name. But often we have cases where the same name has been used more than once for different taxa. For example, here is what ION has for the name "Nystactes".
If I want to map these names to GBIF then these are corresponding taxa with the name "Nystactes":
Clearly the names are almost identical, but there are enough little differences (presence or absence of comma, "o" versus "ö") to make things interesting. To make the mapping I construct a bipartite graph where the nodes are taxon names, divided into two sets based on which database they came from. I then connect the nodes of the graph by edges, weighted by how similar the names are. For example, here is the graph for "Nystactes" (displayed using Google images:

I then compute the maximum weighted bipartite matching using a C++ program I wrote. This matching corresponds to the solid lines in the graph above.
In this way we can make a sensible guess as to how names in the two databases relate to one another.
| Nystactes Bohlke | 2735131 |
| Nystactes | 2787598 |
| Nystactes Gloger 1827 | 4888093 |
| Nystactes Kaup 1829 | 4888094 |
If I want to map these names to GBIF then these are corresponding taxa with the name "Nystactes":
| Nystactes Böhlke, 1957 | 2403398 |
| Nystactes Gloger, 1827 | 2475109 |
| Nystactes Kaup, 1829 | 3239722 |
Clearly the names are almost identical, but there are enough little differences (presence or absence of comma, "o" versus "ö") to make things interesting. To make the mapping I construct a bipartite graph where the nodes are taxon names, divided into two sets based on which database they came from. I then connect the nodes of the graph by edges, weighted by how similar the names are. For example, here is the graph for "Nystactes" (displayed using Google images:
I then compute the maximum weighted bipartite matching using a C++ program I wrote. This matching corresponds to the solid lines in the graph above.
In this way we can make a sensible guess as to how names in the two databases relate to one another.
Tuesday, March 19, 2013
BioNames update - API documentation

One of the fun things about developing web sites is learning new tricks, tools, and techniques. Typically I hack away on my MacBook, and when something seems vaguely usable I stick it on a web server. For BioNames things need to be a little more formalised, especially as I'm collaborating with another developer (Ryan Schenk). Ryan is focussing on the front end, I'm working on the data (harvesting, cleaning, storing).
In most projects I've worked on the code to talk to the database and the code to display results have been the same, it was ugly but it got things. For this project these two aspects have to be much more cleaning separated so that Ryan and I can work independently. One way to do this is to have a well-defined API that Ryan can develop against. This means I can hide the sometimes messy details of how to communicate with the data, and Ryan doesn't need to worry about how to get access to the data.
Nice idea, but to be workable it requires that the API is documented (if it's just me then the documentation is in my head). Documentation is a pain, and it is easy for it to get out of sync with the code such that what the docs say an API does and what it actually does are two separate things (sound familiar?). What would be great is a tool that enables you to write the API documentation, and make that "live" so that the API output can be tested against. In other words, a tool like apiary.io.
Apiary.io is free, very slick, and comes with GitHUb integration. I've started to document the BioNames API at http://docs.bionames.apiary.io/. These documents are "live" in that you can try out the API and get live results from the BioNames database.
I'm sure this is all old news to real software developers (as opposed to people like me who know just enough to get themselves into trouble), but it's quite liberating to start with the API first before worrying about what the web site will look like.
Monday, March 18, 2013
New look Biodiversity Heritage Library launched
Tomorrow the new & improved #bhlib launches!! ow.ly/iVeZb Explore the changes in our Guide! ow.ly/iVf1W
— BHL (@BioDivLibrary) March 17, 2013
The new look Biodiversity Heritage Library has just launched. It's a complete refresh of the old site, based on the Biodiversity Heritage Library–Australia site. If you want an overview of what's new, BHL have published a guide to the new look site. Congrats to involved in the relaunch.
One of the new features draws on the work I've been doing on BioStor. The new BHL interface adds the notion of "parts" of an item, which you can see under the "Table of Contents" tab. For example, the scanned volume 109 of the Proceedings of the Entomological Society of Washington now displays a list of articles within that volume:

This means you can now jump to individual articles. Before you had to scroll through the scan, or click through page numbers until you found what you were after. The screenshot above shows the article "Three new species of chewing lice (Phthiraptera: Ischnocera: Philopteridae) from australian parrots (Psittaciformes: Psittacidae)". The details of this article have been extracted from BioStor, where this article appears as http://biostor.org/reference/55323. You can go directly to this article in BHL using the link http://www.biodiversitylibrary.org/part/69723. As an aside, I've chosen this article because it helps demonstrate that BHL has modern content as well as pre-1923 literature, and this article names a louse, Neopsittaconirmus vincesmithi after a former student of mine, Vince Smith. You're nobody in this field unless you've had a louse named after you ;)
BioStor has over 90,000 articles, but this is a tiny fraction of the articles contained in BHL content, so there's a long way to go until the entire archive is indexed to article level. There will also be errors in the article metadata derived from BioStor. If we invoke Linus's Law ("given enough eyeballs, all bugs are shallow") then having this content in BHL should help expose those errors more rapidly.
As always, I have a few niggles about the site, but I'll save those for another time. For noe, I'm happy to celebrate an extraordinary, open access archive of over 40 million pages. BHL represents one of the few truly indispensable biodiversity resources online.
Friday, March 15, 2013
BioNames ideas - automatically finding synonyms from the literature
One of the biggest pains (and self-inflicted wounds) in taxonomy is synonymy, the existence of multiple names for the same taxon. A common cause of synonymy is moving species to different genera in order to have their name reflect their classification. The consequence of this is any attempt to search the literature for basic biological data runs into the problem that observations published at different times by different researchers (e.g., taxonomists, ecologists, parasitologists) may use different names for the same taxon.
Existing taxonomic databases often have lists of synonyms, but these are incomplete, and typically don't provide any evidence why two names are synonyms.
Reading literature extracted form the Biodiversity Heritage Library I'm struck by how often I come across papers such as taxonomic revisions, museum catalogues, and checklists, that list two names as synonyms. Wouldn't it be great if we could mine these to automatically build lists of synonyms?
One quick and dirty way to do this is look for sets of names that have the same species name but different generic names, e.g.
If such names appear on the same page (i.e., in close proximity) there's a reasonable chance they are synonyms. So, one of the features I'm building in BioNames is an index of names like this. Hence, if we are displaying a page for the name Atlantoxerus getulus that page could also display Sciurus getulus and Xerus getulus as possible synonyms.
There's a lot more that could be done with this sort of approach. For example, this approach only works if the the species name remains unchanged. To improve it we'd need to do things like handle changes to the ending of a species name to agree with the gender of the genus, and cases where the taxa are demoted to subspecies (or promoted to species).
If we were even clever we'd attempt to parse synonymy lists to extract even more synonyms (for an example see Huber and Klump (PDF available here):
Then there's the broader topic of looking at co-occurrence of taxonomic names in general. As I noted a while ago there are examples of pages in BHL that lists taxonomically unrelated taxa that are ecologically closely associated (e.g., hosts and parasites). Hence we could imagine automatically building host-parasite databases by mining the literature. Initially we could simply display lists of names that co-occur frequently. Ideally we'd filter out "accidental" co-occurrences, such as indexes or tables of contents, but there seems to be a lot of potential in automating the extraction of basic information from the taxonomic literature.
Existing taxonomic databases often have lists of synonyms, but these are incomplete, and typically don't provide any evidence why two names are synonyms.
Reading literature extracted form the Biodiversity Heritage Library I'm struck by how often I come across papers such as taxonomic revisions, museum catalogues, and checklists, that list two names as synonyms. Wouldn't it be great if we could mine these to automatically build lists of synonyms?
One quick and dirty way to do this is look for sets of names that have the same species name but different generic names, e.g.
- Atlantoxerus getulus
- Sciurus getulus
- Xerus getulus
If such names appear on the same page (i.e., in close proximity) there's a reasonable chance they are synonyms. So, one of the features I'm building in BioNames is an index of names like this. Hence, if we are displaying a page for the name Atlantoxerus getulus that page could also display Sciurus getulus and Xerus getulus as possible synonyms.
There's a lot more that could be done with this sort of approach. For example, this approach only works if the the species name remains unchanged. To improve it we'd need to do things like handle changes to the ending of a species name to agree with the gender of the genus, and cases where the taxa are demoted to subspecies (or promoted to species).
If we were even clever we'd attempt to parse synonymy lists to extract even more synonyms (for an example see Huber and Klump (PDF available here):
Huber, R., & Klump, J. (2009). Charting taxonomic knowledge through ontologies and ranking algorithms. Computers & Geosciences, 35(4), 862–868. doi:10.1016/j.cageo.2008.02.016
Then there's the broader topic of looking at co-occurrence of taxonomic names in general. As I noted a while ago there are examples of pages in BHL that lists taxonomically unrelated taxa that are ecologically closely associated (e.g., hosts and parasites). Hence we could imagine automatically building host-parasite databases by mining the literature. Initially we could simply display lists of names that co-occur frequently. Ideally we'd filter out "accidental" co-occurrences, such as indexes or tables of contents, but there seems to be a lot of potential in automating the extraction of basic information from the taxonomic literature.
Labels:
BioNames,
co-occurrence,
synonymy
BioNames: yet another taxonomic database
Yet another taxonomic database, this time I can't blame anyone else because I'm the one building it (with some help, as I'll explain below).
BioNames was my entry in EOL's Computable Data Challenge (you can see the proposal here: http://dx.doi.org/10.6084/m9.figshare.92091). In that proposal I outlined my goal:
The bulk of the funding from EOL is going into interface work by Ryan Schenk (@ryanschenk), author of synynyms among other cool things. EOL's Chief Scientist Cyndy Parr (@cydparr) is providing adult supervision ("Chief Scientist", why can't I have a title like that?).
Development of BioNames is taking place in the open as much as we can, so there are some places you can see things unfold:
I've lots of terrible code scattered around which I am in the process of organising into something usable, which I'll then post on GitHub. Working with Ryan is forcing me to be a lot more thoughtful about coding this project, which is a good thing. Currently I'm focussing on building an API that will support the kinds of things we want to do. I'm hoping to make this public shortly.
The original proposal was a tad ambitious (no, really). Most of what I hope to do exists in one form or another, but making it robust and usable is a whole other matter.
As the project takes shape I hope to post updates here. If you have any suggestions feel free to make them. The current target is to have this "out the door" by the end of May.
BioNames was my entry in EOL's Computable Data Challenge (you can see the proposal here: http://dx.doi.org/10.6084/m9.figshare.92091). In that proposal I outlined my goal:
BioNames aims to create a biodiversity “dashboard” where at a glance we can see a summary of the taxonomic and phylogenetic information we have for a given taxon, and that information is seamlessly linked together in one place. It combines classifications from EOL with animal taxonomic names from ION, and bibliographic data from multiple sources including BHL, CrossRef, and Mendeley. The goal is to create a database where the user can drill down from a taxonomic name to see the original description, track the fate of that name through successive revisions, and see other related literature. Publications that are freely available will displayed in situ. If the taxon has been sequenced, the user can see one or more phylogenetic trees for those sequences, where each sequence is in turn linked to the publication that made those sequences available. For a biologist the site provides a quick answer to the basic question “what is this taxon?”, coupled with with graphical displays of the relevant bibliographic and genomic information.
The bulk of the funding from EOL is going into interface work by Ryan Schenk (@ryanschenk), author of synynyms among other cool things. EOL's Chief Scientist Cyndy Parr (@cydparr) is providing adult supervision ("Chief Scientist", why can't I have a title like that?).
Development of BioNames is taking place in the open as much as we can, so there are some places you can see things unfold:
- Key features and milestones are on Trello
- Design details are on GitHub
- Database is hosted by Cloudant
- There is a (currently private) design document in Google Docs. I've posted a snapshot on FigShare (http://dx.doi.org/10.6084/m9.figshare.652203
I've lots of terrible code scattered around which I am in the process of organising into something usable, which I'll then post on GitHub. Working with Ryan is forcing me to be a lot more thoughtful about coding this project, which is a good thing. Currently I'm focussing on building an API that will support the kinds of things we want to do. I'm hoping to make this public shortly.
The original proposal was a tad ambitious (no, really). Most of what I hope to do exists in one form or another, but making it robust and usable is a whole other matter.
As the project takes shape I hope to post updates here. If you have any suggestions feel free to make them. The current target is to have this "out the door" by the end of May.
Wednesday, March 13, 2013
In defence of OpenURL: making bibliographic metadata hackable
This is not a post I'd thought I'd write, because OpenURL is an awful spec. But last week I ended up in vigorous debate on Twitter after I posted what I thought was a casual remark:
This ended up being a marathon thread about OpenURL, accessibility, bibliographic metadata, and more. It spilled over onto a previous blog post (Tight versus loose coupling) where Ed Summers and I debated the merits of Context Object in Span (COinS).
This debate still nags at me because I think there's an underlying assumption that people making bibliographic web sites know what's best for their users.
Ed wrote:
That's fine, I like embedded metadata, both as a consumer and as a provider (I provide Google Scholar-compatible metadata in BioStor). What I object to is the idea that this is all we need to do. Embedded metadata is great if you want to make individual articles visible to search engines:

Tools like Google (or bibliographic managers like Mendeley and Zotero) can "read" the web page, extract structured data, and do something with that. Nice for search engines, nice for repositories (metadata becomes part of their search engine optimisation strategy).
But this isn't the only thing a user might want to do. I often find myself confronted with a list of articles on a web site (e.g., a bibliography on a topic, a list of references cited in a paper, the results of a bibliographic search) and those references have no links. Often those links may not have existed when original web page was published, but may exist now. I'd like a tool that helped me find those links.
If a web site doesn't provide the functionality you need then, luckily, you are not entirely at the mercy of the people who made the decisions about what you can and can't do. Tools like Greasemonkey pioneered the idea that we can hack a web page to make it more useful. I see COinS as an example of this approach. If the web page doesn't provide links, but has embedded COinS then I can use those to create OpenURL links to try and locate those references. I am no longer bound by the limitations of the web page itself.

This strikes me as very powerful, and I use COinS a lot where they are available. For example, CrossRef's excellent search engine supports COinS, which means I can find a reference using that tool, then use the embedded COinS to see whether there is a version of that article digitised by the Biodiversity Heritage Library. This enables me to do stuff that CrossRef itself hasn't anticipated, and that makes their search engine much more valuable to me. In a way this is ironic because CrossRef is predicated on the idea that there is one definitive link to a reference, the DOI.
So, what I found frustrating about the conversation with Ed was that it seemed to me that his insistence on following certain standards was at the expense of functionality that I found useful. If the client is the search engine, or the repository, then COinS do indeed seem to offer little apart from God-awful HTML messing up the page. But if you include the user and accept that users may want to do stuff that you don't (indeed can't) anticipate then COinS are useful. This is the "genius of and", why not support both approaches?
Now, COinS are not the only way to implement what I want to do, we could imagine other ways to do this. But to support the functionality that they offer we need a way to encode metadata in a web page, a way to extract that metadata and form a query URL, and a set of services that know what to do with that URL. OpenURL and COinS provide all of this right now and work. I'd be all for alternative tools that did this more simply than the Byzantine syntax of OpenURL, but in the absence of such tools I stick by my original tweet:
If you publish bibliographic data and don't use COinS ocoins.info you are doing it wrong (I'm looking at you @europepmc_news)
— Roderic Page (@rdmpage) March 8, 2013
This ended up being a marathon thread about OpenURL, accessibility, bibliographic metadata, and more. It spilled over onto a previous blog post (Tight versus loose coupling) where Ed Summers and I debated the merits of Context Object in Span (COinS).
This debate still nags at me because I think there's an underlying assumption that people making bibliographic web sites know what's best for their users.
Ed wrote:
I prefer to encourage publishers to use HTML's metadata facilities using the <meta> tag and microdata/RDFa, and build actually useful tools that do something useful with it, like Zotero or Mendeley have done.
That's fine, I like embedded metadata, both as a consumer and as a provider (I provide Google Scholar-compatible metadata in BioStor). What I object to is the idea that this is all we need to do. Embedded metadata is great if you want to make individual articles visible to search engines:

Tools like Google (or bibliographic managers like Mendeley and Zotero) can "read" the web page, extract structured data, and do something with that. Nice for search engines, nice for repositories (metadata becomes part of their search engine optimisation strategy).
But this isn't the only thing a user might want to do. I often find myself confronted with a list of articles on a web site (e.g., a bibliography on a topic, a list of references cited in a paper, the results of a bibliographic search) and those references have no links. Often those links may not have existed when original web page was published, but may exist now. I'd like a tool that helped me find those links.
If a web site doesn't provide the functionality you need then, luckily, you are not entirely at the mercy of the people who made the decisions about what you can and can't do. Tools like Greasemonkey pioneered the idea that we can hack a web page to make it more useful. I see COinS as an example of this approach. If the web page doesn't provide links, but has embedded COinS then I can use those to create OpenURL links to try and locate those references. I am no longer bound by the limitations of the web page itself.

This strikes me as very powerful, and I use COinS a lot where they are available. For example, CrossRef's excellent search engine supports COinS, which means I can find a reference using that tool, then use the embedded COinS to see whether there is a version of that article digitised by the Biodiversity Heritage Library. This enables me to do stuff that CrossRef itself hasn't anticipated, and that makes their search engine much more valuable to me. In a way this is ironic because CrossRef is predicated on the idea that there is one definitive link to a reference, the DOI.
So, what I found frustrating about the conversation with Ed was that it seemed to me that his insistence on following certain standards was at the expense of functionality that I found useful. If the client is the search engine, or the repository, then COinS do indeed seem to offer little apart from God-awful HTML messing up the page. But if you include the user and accept that users may want to do stuff that you don't (indeed can't) anticipate then COinS are useful. This is the "genius of and", why not support both approaches?
Now, COinS are not the only way to implement what I want to do, we could imagine other ways to do this. But to support the functionality that they offer we need a way to encode metadata in a web page, a way to extract that metadata and form a query URL, and a set of services that know what to do with that URL. OpenURL and COinS provide all of this right now and work. I'd be all for alternative tools that did this more simply than the Byzantine syntax of OpenURL, but in the absence of such tools I stick by my original tweet:
If you publish bibliographic data and don't use COinS you are doing it wrong
Bibliographic metadata pollution
I spend a lot of time searching the web for bibliographic metadata and links to digitised versions of publications. Sometimes I search Google and get nothing, sometimes I get the article I'm after, but often I get something like this:
If I search for Die cestoden der Vogel in Google I get masses of hits for the same thing from multiple sources (e.g., Google Books, Amazon, other booksellers, etc.). For this query we can happily click through pages and pages of results that are all, in some sense, the same thing. Sometimes I get the similar results when searching for an article, multiple hits from sites with metadata on that article, but few, if any with an actual link to the article itself.
One byproduct of putting bibliographic metadata on the web is that we are starting to pollute web space with repetitions of the same (or closely similar) metadata. This makes searching for definitive metadata difficult, never mind actually finding the content itself. In some cases we can use tools such as Google Scholar, which clusters multiple versions of the same reference, but Google Scholar is often poor for the kind of literature I am after (e.g., older taxonomic publications).
As Alan Ruttenberg (@alanruttenbergpoints out, books would seem to be a case where Google could extend its knowledge graph and cluster the books together (using ISBNs, title matching, etc.). But meantime if you think simply pumping out bibliographic metadata is a good thing, spare a thought for those of us trying to wade through the metadata soup looking for the "good stuff".

If I search for Die cestoden der Vogel in Google I get masses of hits for the same thing from multiple sources (e.g., Google Books, Amazon, other booksellers, etc.). For this query we can happily click through pages and pages of results that are all, in some sense, the same thing. Sometimes I get the similar results when searching for an article, multiple hits from sites with metadata on that article, but few, if any with an actual link to the article itself.
One byproduct of putting bibliographic metadata on the web is that we are starting to pollute web space with repetitions of the same (or closely similar) metadata. This makes searching for definitive metadata difficult, never mind actually finding the content itself. In some cases we can use tools such as Google Scholar, which clusters multiple versions of the same reference, but Google Scholar is often poor for the kind of literature I am after (e.g., older taxonomic publications).
As Alan Ruttenberg (@alanruttenbergpoints out, books would seem to be a case where Google could extend its knowledge graph and cluster the books together (using ISBNs, title matching, etc.). But meantime if you think simply pumping out bibliographic metadata is a good thing, spare a thought for those of us trying to wade through the metadata soup looking for the "good stuff".
Friday, March 08, 2013
On Names Attribution, Rights, and Licensing of taxonomic names
Few things have annoyed be as much as the following post on TAXACOM:
I'm trying to work out why this seemingly innocuous post made me so mad. I think this is because I think this fundamentally framing the question the wrong way. Surely the goal is to have a list of names that is global in scope, well documented, and freely usable by all without restriction? Surely we want open and free access to fundamental biodiversity data? In which case, can we please stop having meetings and get on with making this so?
If you frame the discussion as one of "Attribution, Rights and Licensing of names and compilations of names" then you've already lost sight of the prize. You've focussed on the presumed "rights" of name compilers instead.
I would argue that names compilations are somewhat overvalued. They are basically lists of names, sometimes (all to rarely) with some degree of provenance (e.g., a citation to the original use of the name). As I've documented before (e.g., More fictional taxa and the myth of the expert taxonomic database and Fictional taxa) entirely fictional can end up in taxonomic databases with alarming ease. So any claims that these are expert-curated lists should be taken with a pinch of salt.
Furthermore, it is increasingly easy to automate building these lists, given that we have tools for finding names in text, and an ever expanding volume of digitised text becoming available. Indeed, in an ideal world where all taxonomic literature was digitised much of the rationale for taxonomic name databases would disappear (in the same way that library card catalogues are irrelevant in the age of Google). We are fast approaching the point where we can do better than experts. To give just one example, in a recent BHL interview with Gary Poore it was stated that:
A quick check of Google Ngrams shows this to be simply false:
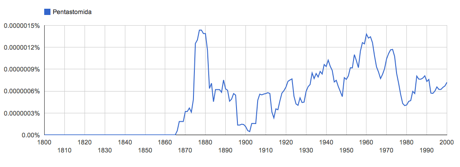
I don't need taxonomic expertise to see this, I simply need decent text indexing. So, if you have a list of names, you have something that it will soon be largely possible to recreate using automated methods (i.e., text mining). With a little sophistication we could mine the literature for further details, such as synonymy, etc. Annotation and clarification of a few "edge cases" where things get tricky will always be needed, but if you want to argue that your lists deserves "Attribution, Rights and Licensing" then you fail to realise that your list is going to be increasing easy to recreate simply by crawling the web.
It seems to me that most taxonomic databases are little more than digitised 5x3 index cards, and lack any details on the provenance of the names they contain. They often don't have links to the primary literature, and if they do cite that literature they typically do so in a way that makes it hard to find the actual publication. I once gave a talk which included the slide below showing taxonomic databases as being "in the way" between taxonomists and users of taxonomic information:

In the old days building taxonomic databases required expertise and access to obscure, hard to find, physical literature. A catalogue of names was a way to summarise that information (since we couldn't share access). Now we are in an age where more and more primary taxonomic information is available to all, which removes most of the rationale for taxonomic databases. Users can go directly to taxonomic information themselves, which mean they can get the "good stuff", and maybe even cite it (giving us provenance and credit, which I regard as basically the same thing). In many ways taxonomic databases are transitional phenomena (like phone directories, remember those), and one could argue are now in the way of the taxonomists' Holy Grail, getting their work cited.
Lastly, any discussion of "Attribution, Rights and Licensing of names and compilations of names" reflects one of the great self inflicted wounds of biodiversity informatics, namely the reluctance to freely share data. As we speak terrabytes of genomics data are whizzing around the planet, people are downloading entire copies of GenBank and creating new databases. All of this without people fussing over "Attribution, Rights and Licensing." It's time for taxonomic databases to get over themselves and focus on making biodiversity data as accessible and available as genomics data.
The Global Names project will host a workshop to explore options and to make recommendations as to issues that relate to Attribution, Rights and Licensing of names and compilations of names. The aim of the workshop is a report that clarifies if and how we share names.
We seek submissions from all interested parties - nomenclaturalists, taxonomists, aggregators, and users of names. Let us know what (you think) intellectual property rights apply or what rights should be associated with names and compilations of names. How can those who compile names get useful attribution for names, and what responsibilities do they have to ensure that information is authoritative. If there are rights, what kind of licensing is appropriate.
Contributions can be submitted http://names-attribution-rights-and-licensing.wikia.com/wiki/Main_Page, where you will find more information about this event.
I'm trying to work out why this seemingly innocuous post made me so mad. I think this is because I think this fundamentally framing the question the wrong way. Surely the goal is to have a list of names that is global in scope, well documented, and freely usable by all without restriction? Surely we want open and free access to fundamental biodiversity data? In which case, can we please stop having meetings and get on with making this so?
If you frame the discussion as one of "Attribution, Rights and Licensing of names and compilations of names" then you've already lost sight of the prize. You've focussed on the presumed "rights" of name compilers instead.
I would argue that names compilations are somewhat overvalued. They are basically lists of names, sometimes (all to rarely) with some degree of provenance (e.g., a citation to the original use of the name). As I've documented before (e.g., More fictional taxa and the myth of the expert taxonomic database and Fictional taxa) entirely fictional can end up in taxonomic databases with alarming ease. So any claims that these are expert-curated lists should be taken with a pinch of salt.
Furthermore, it is increasingly easy to automate building these lists, given that we have tools for finding names in text, and an ever expanding volume of digitised text becoming available. Indeed, in an ideal world where all taxonomic literature was digitised much of the rationale for taxonomic name databases would disappear (in the same way that library card catalogues are irrelevant in the age of Google). We are fast approaching the point where we can do better than experts. To give just one example, in a recent BHL interview with Gary Poore it was stated that:
For example, the name widely used name Pentastomida itself was widely attributed to Diesing, 1836, but the word did not appear in the literature until 1905.
A quick check of Google Ngrams shows this to be simply false:

I don't need taxonomic expertise to see this, I simply need decent text indexing. So, if you have a list of names, you have something that it will soon be largely possible to recreate using automated methods (i.e., text mining). With a little sophistication we could mine the literature for further details, such as synonymy, etc. Annotation and clarification of a few "edge cases" where things get tricky will always be needed, but if you want to argue that your lists deserves "Attribution, Rights and Licensing" then you fail to realise that your list is going to be increasing easy to recreate simply by crawling the web.
It seems to me that most taxonomic databases are little more than digitised 5x3 index cards, and lack any details on the provenance of the names they contain. They often don't have links to the primary literature, and if they do cite that literature they typically do so in a way that makes it hard to find the actual publication. I once gave a talk which included the slide below showing taxonomic databases as being "in the way" between taxonomists and users of taxonomic information:

In the old days building taxonomic databases required expertise and access to obscure, hard to find, physical literature. A catalogue of names was a way to summarise that information (since we couldn't share access). Now we are in an age where more and more primary taxonomic information is available to all, which removes most of the rationale for taxonomic databases. Users can go directly to taxonomic information themselves, which mean they can get the "good stuff", and maybe even cite it (giving us provenance and credit, which I regard as basically the same thing). In many ways taxonomic databases are transitional phenomena (like phone directories, remember those), and one could argue are now in the way of the taxonomists' Holy Grail, getting their work cited.
Lastly, any discussion of "Attribution, Rights and Licensing of names and compilations of names" reflects one of the great self inflicted wounds of biodiversity informatics, namely the reluctance to freely share data. As we speak terrabytes of genomics data are whizzing around the planet, people are downloading entire copies of GenBank and creating new databases. All of this without people fussing over "Attribution, Rights and Licensing." It's time for taxonomic databases to get over themselves and focus on making biodiversity data as accessible and available as genomics data.
Friday, March 01, 2013
Why the ICZN is in trouble
There are many reasons why the International Commission on Zoological Nomenclature (ICZN) is in trouble, but fundamentally I think it's because of situation illustrated by following diagram.

Based on an analysis of the Index of Organism Names (ION) database that I'm currently working on, there are around 3.8 million animal names (I define "animal" loosely, the ICZN covers a number of eukaryote groups), of which around 1.5 million are "original combinations", that is, the name as originally published. The other 2 million plus names are synonyms, spelling variations, etc.
Of these 3.8 million names the ICZN itself can say very little. It has placed some 12,600 names (around 0.3% of the total) on its Official Lists and Indexes (which is where it records decisions on nomenclature), and its new register of names, ZooBank, has less than 100,000 names (i.e., less than 3% of all animal names).
The ICZN doesn't have a comprehensive database of animal names, so it can't answer the most basic questions one might have about names (e.g., "is this a name?", "can I use this name, or has somebody already used it?", "what other names have people used for this taxon?", "where was this name originally published?", "can I see the original description?", "who first said these two names are synonyms?", and so on). The ICZN has no answer to these questions. In the absence of these services, it is reduced to making decisions about a tiny fraction of the names that are in use (and there is no database of these decisions). It is no wonder that it is in such trouble.

Based on an analysis of the Index of Organism Names (ION) database that I'm currently working on, there are around 3.8 million animal names (I define "animal" loosely, the ICZN covers a number of eukaryote groups), of which around 1.5 million are "original combinations", that is, the name as originally published. The other 2 million plus names are synonyms, spelling variations, etc.
Of these 3.8 million names the ICZN itself can say very little. It has placed some 12,600 names (around 0.3% of the total) on its Official Lists and Indexes (which is where it records decisions on nomenclature), and its new register of names, ZooBank, has less than 100,000 names (i.e., less than 3% of all animal names).
The ICZN doesn't have a comprehensive database of animal names, so it can't answer the most basic questions one might have about names (e.g., "is this a name?", "can I use this name, or has somebody already used it?", "what other names have people used for this taxon?", "where was this name originally published?", "can I see the original description?", "who first said these two names are synonyms?", and so on). The ICZN has no answer to these questions. In the absence of these services, it is reduced to making decisions about a tiny fraction of the names that are in use (and there is no database of these decisions). It is no wonder that it is in such trouble.
Subscribe to:
Comments (Atom)
