It's slowly
dawning on me that many of the ingredients for an alternative different way to browse scientific articles may already be in place. After my first
crude efforts at what an iPad reader might look like I've started afresh with a new attempt, based on the
Sencha Touch framework. The goal here isn't to make a polished app, but rather to get a sense of what could be done.
The first goal is to be able to browse the literature as if it was a connected series of documents (which is what, of course, it is). This requires taking the full text of an article, extracting the citations, and making them links to further documents (also with their citations extracted, and so on). Leaving aside the obvious problem that this approach is limited to open access articles, an app that does this is going to have to store a lot of bibliographic data as the reader browses the literature (otherwise we going to have to do all the processing on the fly, and that's not going to be fast enough). So, we need some storage.
MySQLOne option is to write a MySQL database to hold articles, books, etc. Doable (I've done more of these than I care to remember), but things get messy pretty quickly, especially as you add functionality (tags, fulltext, figures, etc.).
RDFAnother option is to use RDF and a tripe store. I've played with linked data quite a bit lately (see previous "Friday follies"
here and
here), and I thought that a triple store would be a great way support an article browser (especially as we add additional kinds of data, such as sequences, specimens, phylogenies, etc.). But linked data is a mess. For the things I care about there are either no canonical identifiers, or too many, and rarely does the primary data provider served linked data compliant URLs (e.g., NCBI), hence we end up with a plethora of wrappers around these sources. Then there's the issue of what vocabularies to use (once again, there are either none, or too many). As a query language SPARQL isn't great, and don't even get me started on the issue of editing data. OK, so I get the whole idea of linked data, it's just that the overhead of getting anything done seems too high. You've got to get a lot of ducks to line up.
CounchDB
So, I started playing with
CounchDB, in a fairly idle way. I'd had a look before, but didn't really get my head around the very different way of querying a database that CouchDB requires. Despite this learning curve, CouchDB has some great features. It stores documents in JSON, which makes it trivial to add data as objects (instead of mucking around with breaking them up into tables for SQL, or atomising them into triples for RDF), it supports versioning right out of the box (vital because metadata is often wrong and needs to be tidied up), and you talk to it using HTTP, which means no middleware to get in the way. You just point your browser (or curl, or whatever HTTP tool you have) and send GET, POST, PUT, or DELETE commands. And now it's
in the cloud.
In some ways ending up with CouchDB (or something similar) seems inevitable. The one "semantic web" tool that I've made most use of is
Semantic MediaWiki, which powers the
NCBI to Wikipedia mapping I created in June. Semantic Mediawiki has it's uses, but occasionally it has
driven me to distraction. But, when you get down to it, Semantic Mediawiki is really just a versioned document store (where the documents are typically key-value pairs), over which have been laid a pretty limited query language and some RDF export features. Put like this, most of the huge Mediawiki engine underlying Semantic MediaWiki isn't needed, so why not cut to the chase and use a purpose-built versioned document store? Enter CouchDB.
Browsing and Mendeley So, what I have in mind is a browser that crawls a document, extracting citations, and enabling the reader to explore those. Eventually it will also extract all the other chocolatey goodness in an article (sequences, specimens, taxonomic names, etc.), but for now I'm focussing on articles and citations. A browser would need to store article metadata (say, each time it encounters an article for the first time), as well as update existing metadata (by adding missing DOIs, PubMed ids, citations, etc.), so what easier way than as JSON in a document store such as CouchDB? This is what I'm exploring at the moment, but let's take a step back for a second.
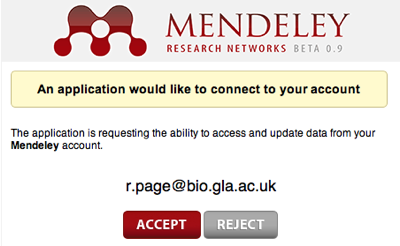
The
Mendeley API, as
poorly developed as it is, could be treated as essentially a wrapper around a JSON document store (the API stores and returns JSON), and
speaks HTTP. So, we could imagine a browser that crawls the Mendeley database, adding papers that aren't in Mendeley as it goes. The act of browsing and reading would actively contribute to the database. Of course, we could spin this around, and argue that a crawler + CouchDB could pretty effectively create a clone of Mendeley's database (albeit without the social networking features that come with have a large user community).
This is another reason why the current crop of iPad article viewers,
Mendeley's included, are so disappointing. There's the potential to completely change the way we interact with the scientific literature (instead of passively consuming PDFs), and Mendeley is ideally positioned to support this. Yes, I realise that for the vast majority of people being able to manage their PDFs and format bibliographies in MS Word are
the killer features, but, seriously, is that all we aspire too?

 Yesterday I uploaded a manuscript to Nature Precedings that describes the inner workings of
Yesterday I uploaded a manuscript to Nature Precedings that describes the inner workings of 



 In previous articles I've looked at how various apps display scientific articles. The apps I looked at were:
In previous articles I've looked at how various apps display scientific articles. The apps I looked at were:
