TL;DR
The Data Citation Corpus is still riddled with errors, and it is unclear to what extent it measures citation (resuse of data) versus publication (are most citations between the data and the original publication)?
These are some brief notes on the latest version (v. 2) of the Data Citation Corpus, relased shortly before the Make Data Count Summit 2024, which also included a discussion on the practical uses of the corpus.
I downloaded version 2 from Zenodo doi:10.5281/zenodo.13376773. The data is in JSON format, which I then loaded into CouchDB to play with. Loading the data was relatively quick using CouchDB’s “bulk upload” feature, although building indexes to explore the data takes a little while.
What follows is a series of charts constructed using Vega-Lite.
The top 20 repositories
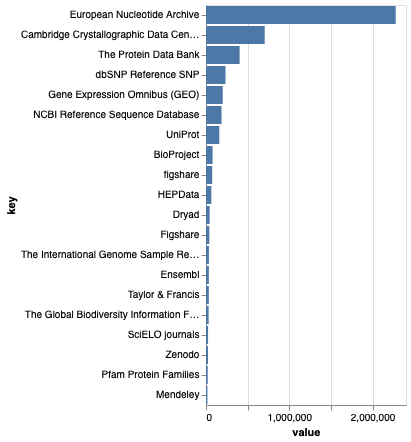
The chart above shows the top 20 repositories by number of citations. The biggest repository by some distance is the European Nucleotide Archive, so most of the data being cited are DNA sequences. Those working on biodiversity might be plessed to see that GBIF is 16th out of 20.
Note that Figshare appears twice, as “Figshare” and “figshare”, so there are problems with data cleaning. It’s actually worse than this, because the repository “Taylor & Francis” is an branded instance of Figshare: https://tandf.figshare.com. So if we want to measure the impact of the Figshare repository we will need to cluster the different spellings, as well as check the DOIs for each repository (T&F data DOIs still carry Figshare branding, e.g.doi:10.6084/m9.figshare.14103264.
The vast majority of data in the “Taylor & Francis” is published by Informa UK Ltd which is the parent company of Taylor & Francis. If you visit an article in an Informa journal, such as Enhanced Selectivity of Ultraviolet-Visible Absorption Spectroscopy with Trilinear Decomposition on Spectral pH Measurements for the Interference-Free Determination of Rutin and Isorhamnetin in Chinese Herbal Medicine the supplementary information is stored in Figshare. These links between the publication and the supplementary information are treated as “citations” in the corpus. Is this what we mean by citation? If so, we are not measuring the use of data, but rather its publication.
Top 20 publishers
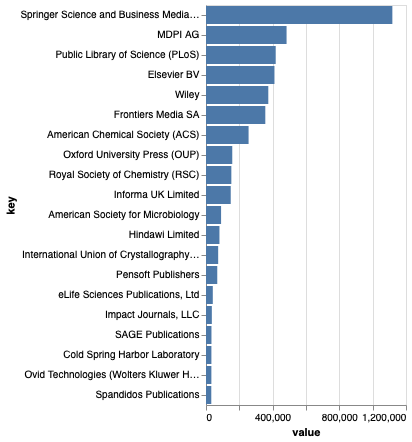
The top 20 publishers of articles that cite data in the corpus are shown above. It would be interesting to know how much this reflects publication policies of these publishers (e.g., open versus closed access, indexing in Pubmed, availability of XML, etc.) versus actual citation of data. Biodiversity people might be pleased to see Pensoft appearing in the top 20.
GBIF
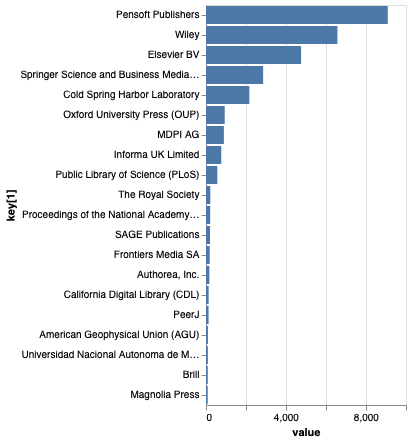
You can also explore the data by individual repository. For example, the top 20 publishers of articles citing data in GBIF shows Pensoft at number one. This reflects the subject matter of Pensoft journals, and Pensoft’s focus on best practices for publishing data. From GBIF’s perspective, perhaps that organisation would want to extend their reach beyond core biodiversity journals.
Citation
The vast majority of data in the Data Citation Corpus is cited only once.
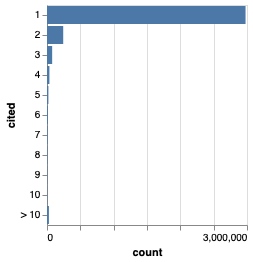
Given that much of these “citations” may be by the publication that makes the data available, it’s not clear to me that the corpus is actually measuring citation (i.e., reuse of the data). Instead it may just be measuring publication (e.g., the link betwene a paper and its supplementary data). To answer this we’d need to drill down into the data more.
Note that some data items have large numbers of citations, the highest is “LY294002” with 9983 citations, with the next being “A549” with 5883 citations. LY294002 is a chemical compound that acts as an inhibitor, and A549 is cell type. The citation corpus regards both as accession numbers for sequences(!). Hence it’s likely that the most cited data records are not data at all, but false matches to other entities, such as chemicals and cells. These false matches are still a major problem for the corpus.
Summary
I think there is so much potential here, but there are significant data quality issues. Anyone basing metrics upon this corpus would need to proceed very carefully.
Written with StackEdit.