For a short, but reasonably technical sumary of what I think the issues are, please read this "
Technical Report", which I presented at the Workshop on Database Issues in Biological Databases (
DBiBD) in Edinburgh in January 2005. This document is itself based on a BBSCR grant proposal which was funded. Here is the abstract.
This note outlines some of the key intellectual obstacles that stand in the way of creating a usable phylogenetic database. These challenges include the need to accommodate multiple taxonomic names and classifications, and the need for tools to query trees in biologically meaningful ways. Until these problems are addressed, and a taxonomically intelligent phylogenetic database created, much of our phylogenetic knowledge will languish in the pages of journals.
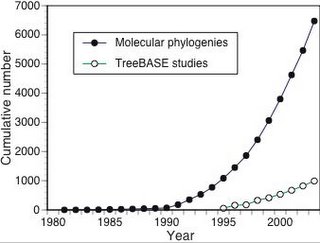
One of my biggest concerns is the growing gap between how many phylogenies are published each year, and those that make it into the best known phylogenetic database we have,
TreeBASE. This graph shows the cumulative growth of publications on phylogenetics, based on the number of papers found in the Web of Science by searching on the key words “molecular” and “phylogenetic” since 1981 the growth of the TreeBASE, which launched in 1996 (a study in TreeBASE is equivalent to a single paper). The idea for this diagram came from Mark Pagel's article "Inferring the historical patterns of biological evolution" (
doi:10.1038/44766).
 One of my biggest concerns is the growing gap between how many phylogenies are published each year, and those that make it into the best known phylogenetic database we have, TreeBASE. This graph shows the cumulative growth of publications on phylogenetics, based on the number of papers found in the Web of Science by searching on the key words “molecular” and “phylogenetic” since 1981 the growth of the TreeBASE, which launched in 1996 (a study in TreeBASE is equivalent to a single paper). The idea for this diagram came from Mark Pagel's article "Inferring the historical patterns of biological evolution" (doi:10.1038/44766).
One of my biggest concerns is the growing gap between how many phylogenies are published each year, and those that make it into the best known phylogenetic database we have, TreeBASE. This graph shows the cumulative growth of publications on phylogenetics, based on the number of papers found in the Web of Science by searching on the key words “molecular” and “phylogenetic” since 1981 the growth of the TreeBASE, which launched in 1996 (a study in TreeBASE is equivalent to a single paper). The idea for this diagram came from Mark Pagel's article "Inferring the historical patterns of biological evolution" (doi:10.1038/44766).
No comments:
Post a Comment
Note: only a member of this blog may post a comment.