The problem addressed by Donat is the issue of multiple strings for the same journal (e.g., the plethora of different abbreviations and permutations people use to refer to the same journal). In trying to make sense of this mess there are a couple of strategies we can use. One is to cluster the strings into sets that we think refer to the same thing, e.g.:
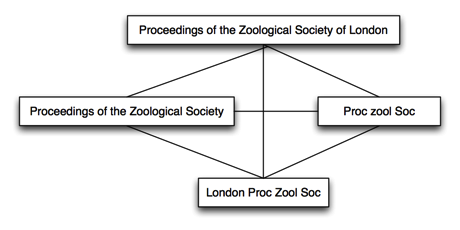
We could then synthesise the preferred journal name from this set. We could make some sort of consensus string, for example. There are also some quite nice Bayesian methods for combining contradictory metadata.
Another approach, which I use, is to map the strings to a third party identifier, in this case an ISSN:
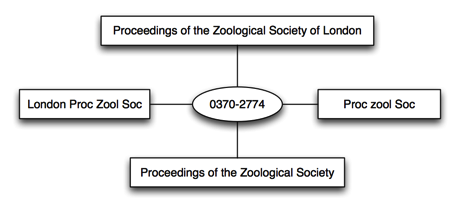
Once I've done this I can use the identifier to refer to the journal, hence ultimately I don't particularly care what string is best for the journal (indeed, I can defer to a third party for this decision).
The point is obsessing with clean, "correct" bibliographic metadata is something of a fool's errand. Obviously, it's nice to have clean metadata if you can get it, but in many cases there is no exact answer to what is the correct metadata. Some journals have multiple names (e.g., in different languages), some run different volume numbering schemes in parallel, and date of publication can be rather problematic (see my Mendeley group on publication dates). If we can map a publication to a globally unique identifier, such as a DOI, then we can sidestep this issue and focus on what I think really matters - linking data together.